Information
- Publication Type: Journal Paper with Conference Talk
- Workgroup(s)/Project(s):
- Date: November 2019
- Journal: Computer Graphics Forum
- Volume: 38
- Number: 7
- Lecturer: Tobias Klein
- Event: Pacific Graphics 2109
- DOI: 10.1111/cgf.13816
- Call for Papers: Call for Paper
- Conference date: 2019
- Pages: 57 – 68
Abstract
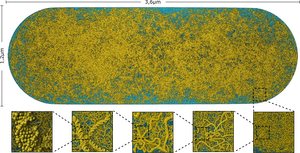
Visualization of biological mesoscale models provides a glimpse at the inner workings of living cells. One of the most complex components of these models is DNA, which is of fundamental importance for all forms of life. Modeling the 3D structure of genomes has previously only been attempted by sequential approaches. We present the first parallel approach for the instant construction of DNA structures. Traditionally, such structures are generated with algorithms like random walk, which have inherent sequential constraints. These algorithms result in the desired structure, are easy to control, and simple to formulate. Their execution, however, is very time-consuming, as they are not designed to exploit parallelism. We propose an approach to parallelize the process, facilitating an implementation on the GPU.Additional Files and Images
Weblinks
BibTeX
@article{klein_2019_PGG,
title = "Parallel Generation and Visualization of Bacterial Genome
Structures",
author = "Tobias Klein and Peter Mindek and Ludovic Autin and David
Goodsell and Arthur Olson and Eduard Gr\"{o}ller and Ivan
Viola",
year = "2019",
abstract = "Visualization of biological mesoscale models provides a
glimpse at the inner workings of living cells. One of the
most complex components of these models is DNA, which is of
fundamental importance for all forms of life. Modeling the
3D structure of genomes has previously only been attempted
by sequential approaches. We present the first parallel
approach for the instant construction of DNA structures.
Traditionally, such structures are generated with algorithms
like random walk, which have inherent sequential
constraints. These algorithms result in the desired
structure, are easy to control, and simple to formulate.
Their execution, however, is very time-consuming, as they
are not designed to exploit parallelism. We propose an
approach to parallelize the process, facilitating an
implementation on the GPU.",
month = nov,
journal = "Computer Graphics Forum",
volume = "38",
number = "7",
doi = "10.1111/cgf.13816",
pages = "57--68",
URL = "https://www.cg.tuwien.ac.at/research/publications/2019/klein_2019_PGG/",
}