Information
- Publication Type: Journal Paper with Conference Talk
- Workgroup(s)/Project(s):
- Date: 2017
- Journal: IEEE Transactions on Visualization and Computer Graphics
- Volume: 24
- Number: 1
- Location: Phoenix, Arizona
- Lecturer: Peter Mindek
- Event: IEEE Vis 2017
- Conference date: 29. September 2017 – 7. October 2017
- Keywords: Biological visualization, remote rendering, public dissemination
Abstract
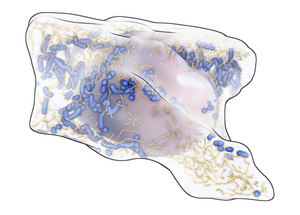
We propose a system to facilitate biology communication by developing a pipeline to support the instructional visualization of heterogeneous biological data on heterogeneous user-devices. Discoveries and concepts in biology are typically summarized with illustrations assembled manually from the interpretation and application of heterogenous data. The creation of such illustrations is time consuming, which makes it incompatible with frequent updates to the measured data as new discoveries are made. Illustrations are typically non-interactive, and when an illustration is updated, it still has to reach the user. Our system is designed to overcome these three obstacles. It supports the integration of heterogeneous datasets, reflecting the knowledge that is gained from different data sources in biology. After pre-processing the datasets, the system transforms them into visual representations as inspired by scientific illustrations. As opposed to traditional scientific illustration these representations are generated in real-time - they are interactive. The code generating the visualizations can be embedded in various software environments. To demonstrate this, we implemented both a desktop application and a remote-rendering server in which the pipeline is embedded. The remote-rendering server supports multi-threaded rendering and it is able to handle multiple users simultaneously. This scalability to different hardware environments, including multi-GPU setups, makes our system useful for efficient public dissemination of biological discoveries.Additional Files and Images
Weblinks
BibTeX
@article{mindek-2017-marion,
title = "Visualization Multi-Pipeline for Communicating Biology",
author = "Peter Mindek and David Kou\v{r}il and Johannes Sorger and
David Toloudis and Blair Lyons and Graham Johnson and Eduard
Gr\"{o}ller and Ivan Viola",
year = "2017",
abstract = "We propose a system to facilitate biology communication by
developing a pipeline to support the instructional
visualization of heterogeneous biological data on
heterogeneous user-devices. Discoveries and concepts in
biology are typically summarized with illustrations
assembled manually from the interpretation and application
of heterogenous data. The creation of such illustrations is
time consuming, which makes it incompatible with frequent
updates to the measured data as new discoveries are made.
Illustrations are typically non-interactive, and when an
illustration is updated, it still has to reach the user. Our
system is designed to overcome these three obstacles. It
supports the integration of heterogeneous datasets,
reflecting the knowledge that is gained from different data
sources in biology. After pre-processing the datasets, the
system transforms them into visual representations as
inspired by scientific illustrations. As opposed to
traditional scientific illustration these representations
are generated in real-time - they are interactive. The code
generating the visualizations can be embedded in various
software environments. To demonstrate this, we implemented
both a desktop application and a remote-rendering server in
which the pipeline is embedded. The remote-rendering server
supports multi-threaded rendering and it is able to handle
multiple users simultaneously. This scalability to different
hardware environments, including multi-GPU setups, makes our
system useful for efficient public dissemination of
biological discoveries. ",
journal = "IEEE Transactions on Visualization and Computer Graphics",
volume = "24",
number = "1",
keywords = "Biological visualization, remote rendering, public
dissemination",
URL = "https://www.cg.tuwien.ac.at/research/publications/2017/mindek-2017-marion/",
}

 Paper
Paper Preview Movie
Preview Movie
