Information
- Publication Type: Journal Paper with Conference Talk
- Workgroup(s)/Project(s):
- Date: January 2019
- Journal: IEEE Transactions on Visualization and Computer Graphics
- Volume: 25
- Open Access: yes
- Note: SciVis Best Paper Honorable Mention
- Location: Berlin, Germany
- Lecturer: David Kouřil
- Event: IEEE VIS
- DOI: 10.1109/TVCG.2018.2864491
- Call for Papers: Call for Paper
- Conference date: November 2018 – March 2018
- Pages: 977 – 986
- Keywords: labeling, multi-scale data, multi-instance data
Abstract
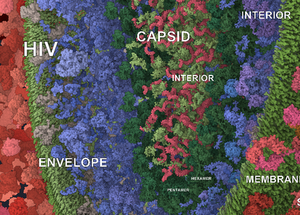
Labeling is intrinsically important for exploring and understanding complex environments and models in a variety of domains. We present a method for interactive labeling of crowded 3D scenes containing very many instances of objects spanning multiple scales in size. In contrast to previous labeling methods, we target cases where many instances of dozens of types are present and where the hierarchical structure of the objects in the scene presents an opportunity to choose the most suitable level for each placed label. Our solution builds on and goes beyond labeling techniques in medical 3D visualization, cartography, and biological illustrations from books and prints. In contrast to these techniques, the main characteristics of our new technique are: 1) a novel way of labeling objects as part of a bigger structure when appropriate, 2) visual clutter reduction by labeling only representative instances for each type of an object, and a strategy of selecting those. The appropriate level of label is chosen by analyzing the scene's depth buffer and the scene objects' hierarchy tree. We address the topic of communicating the parent-children relationship between labels by employing visual hierarchy concepts adapted from graphic design. Selecting representative instances considers several criteria tailored to the character of the data and is combined with a greedy optimization approach. We demonstrate the usage of our method with models from mesoscale biology where these two characteristics-multi-scale and multi-instance-are abundant, along with the fact that these scenes are extraordinarily dense.Additional Files and Images
Additional images and videos
Additional files
Weblinks
- Process video
Video showing the progress of the implementation of the method described in the paper. - Conference Talk Recording
Recording of the talk delivered at IEEE VIS 2018 in Berlin - DOI: 10.1109/TVCG.2018.2864491
BibTeX
@article{kouril-2018-LoL,
title = "Labels on Levels: Labeling of Multi-Scale Multi-Instance and
Crowded 3D Biological Environments",
author = "David Kou\v{r}il and Ladislav \v{C}mol\'{i}k and Barbora
Kozlikova and Hsiang-Yun Wu and Graham Johnson and David
Goodsell and Arthur Olson and Eduard Gr\"{o}ller and Ivan
Viola",
year = "2019",
abstract = "Labeling is intrinsically important for exploring and
understanding complex environments and models in a variety
of domains. We present a method for interactive labeling of
crowded 3D scenes containing very many instances of objects
spanning multiple scales in size. In contrast to previous
labeling methods, we target cases where many instances of
dozens of types are present and where the hierarchical
structure of the objects in the scene presents an
opportunity to choose the most suitable level for each
placed label. Our solution builds on and goes beyond
labeling techniques in medical 3D visualization,
cartography, and biological illustrations from books and
prints. In contrast to these techniques, the main
characteristics of our new technique are: 1) a novel way of
labeling objects as part of a bigger structure when
appropriate, 2) visual clutter reduction by labeling only
representative instances for each type of an object, and a
strategy of selecting those. The appropriate level of label
is chosen by analyzing the scene's depth buffer and the
scene objects' hierarchy tree. We address the topic of
communicating the parent-children relationship between
labels by employing visual hierarchy concepts adapted from
graphic design. Selecting representative instances considers
several criteria tailored to the character of the data and
is combined with a greedy optimization approach. We
demonstrate the usage of our method with models from
mesoscale biology where these two
characteristics-multi-scale and multi-instance-are abundant,
along with the fact that these scenes are extraordinarily
dense.",
month = jan,
journal = "IEEE Transactions on Visualization and Computer Graphics",
volume = "25",
note = "SciVis Best Paper Honorable Mention",
doi = "10.1109/TVCG.2018.2864491",
pages = "977--986",
keywords = "labeling, multi-scale data, multi-instance data",
URL = "https://www.cg.tuwien.ac.at/research/publications/2019/kouril-2018-LoL/",
}

 paper
paper

