Information
- Publication Type: Journal Paper (without talk)
- Workgroup(s)/Project(s):
- Date: January 2019
- DOI: 10.1016/j.jmb.2018.09.004.
- Journal: Journal of Molecular Biology
- Number: 31
- Open Access: no
- Volume: 6
- Pages: 1049 – 1070
- Keywords: molecular visualization, molecular dynamics, modelitics, DNA nanotechnology, visual abstraction
Abstract
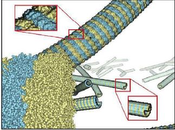
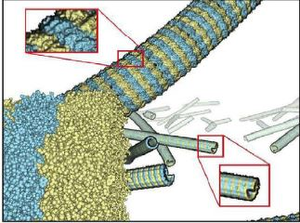
We provide a high-level survey of multiscale molecular visualization techniques, with a focus on application-domain questions, challenges, and tasks. We provide a general introduction to molecular visualization basicsand describe a number of domain-specific tasks that drive this work. These tasks, in turn, serve as the generalstructure of the following survey. First, we discuss methods that support the visual analysis of moleculardynamics simulations. We discuss, in particular, visual abstraction and temporal aggregation. In the secondpart, we survey multiscale approaches that support the design, analysis, and manipulation of DNAnanostructures and related concepts for abstraction, scale transition, scale-dependent modeling, andnavigation of the resulting abstraction spaces. In the third part of the survey, we showcase approaches thatsupport interactive exploration within large structural biology assemblies up to the size of bacterial cells.We describe fundamental rendering techniques as well as approaches for element instantiation, visibilitymanagement, visual guidance, camera control, and support of depth perception. We close the survey with abrief listing of important tools that implement many of the discussed approaches and a conclusion thatprovides some research challenges in the field.
Additional Files and Images
Additional images and videos
Additional files
Weblinks
BibTeX
@article{Miao_2019,
title = "Multiscale Molecular Visualization",
author = "Haichao Miao and Tobias Klein and David Kou\v{r}il and Peter
Mindek and Karsten Schatz and Eduard Gr\"{o}ller and Barbora
Kozlikova and Tobias Isenberg and Ivan Viola",
year = "2019",
abstract = "We provide a high-level survey of multiscale molecular
visualization techniques, with a focus on application-domain
questions, challenges, and tasks. We provide a general
introduction to molecular visualization basicsand describe a
number of domain-specific tasks that drive this work. These
tasks, in turn, serve as the generalstructure of the
following survey. First, we discuss methods that support the
visual analysis of moleculardynamics simulations. We
discuss, in particular, visual abstraction and temporal
aggregation. In the secondpart, we survey multiscale
approaches that support the design, analysis, and
manipulation of DNAnanostructures and related concepts for
abstraction, scale transition, scale-dependent modeling,
andnavigation of the resulting abstraction spaces. In the
third part of the survey, we showcase approaches thatsupport
interactive exploration within large structural biology
assemblies up to the size of bacterial cells.We describe
fundamental rendering techniques as well as approaches for
element instantiation, visibilitymanagement, visual
guidance, camera control, and support of depth perception.
We close the survey with abrief listing of important tools
that implement many of the discussed approaches and a
conclusion thatprovides some research challenges in the
field.",
month = jan,
doi = "10.1016/j.jmb.2018.09.004.",
journal = "Journal of Molecular Biology",
number = "31",
volume = "6",
pages = "1049--1070",
keywords = "molecular visualization, molecular dynamics, modelitics, DNA
nanotechnology, visual abstraction",
URL = "https://www.cg.tuwien.ac.at/research/publications/2019/Miao_2019/",
}

 Paper
Paper