Information
- Publication Type: Conference Paper
- Workgroup(s)/Project(s): not specified
- Date: September 2022
- ISBN: 978-3-03868-177-9
- Publisher: The Eurographics Association
- Open Access: yes
- Location: Wien
- Lecturer: Caroline Magg
- Event: Eurographics Workshop on Visual Computing for Biology and Medicine (VCBM2022)
- DOI: 10.2312/vcbm.20221193
- Booktitle: Eurographics Workshop on Visual Computing for Biology and Medicine (VCBM 2022)
- Pages: 5
- Conference date: 22. September 2022
– 23. September 2022
- Pages: 111 – 115
- Keywords: Visual Analytics, Life and medical sciences, Applied computing
Abstract
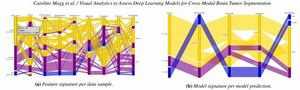
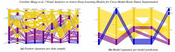
Accurate delineations of anatomically relevant structures are required for cancer treatment planning. Despite its accuracy, manual labeling is time-consuming and tedious-hence, the potential of automatic approaches, such as deep learning models, is being investigated. A promising trend in deep learning tumor segmentation is cross-modal domain adaptation, where knowledge learned on one source distribution (e.g., one modality) is transferred to another distribution. Yet, artificial intelligence (AI) engineers developing such models, need to thoroughly assess the robustness of their approaches, which demands a deep understanding of the model(s) behavior. In this paper, we propose a web-based visual analytics application that supports the visual assessment of the predictive performance of deep learning-based models built for cross-modal brain tumor segmentation. Our application supports the multi-level comparison of multiple models drilling from entire cohorts of patients down to individual slices, facilitates the analysis of the relationship between image-derived features and model performance, and enables the comparative exploration of the predictive outcomes of the models. All this is realized in an interactive interface with multiple linked views. We present three use cases, analyzing differences in deep learning segmentation approaches, the influence of the tumor size, and the relationship of other data set characteristics to the performance. From these scenarios, we discovered that the tumor size, i.e., both volumetric in 3D data and pixel count in 2D data, highly affects the model performance, as samples with small tumors often yield poorer results. Our approach is able to reveal the best algorithms and their optimal configurations to support AI engineers in obtaining more insights for the development of their segmentation models.
Additional Files and Images
Additional images and videos
Additional files
Weblinks
BibTeX
@inproceedings{magg2022,
title = "Visual Analytics to Assess Deep Learning Models for
Cross-Modal Brain Tumor Segmentation",
author = "Caroline Magg and Renata Raidou",
year = "2022",
abstract = "Accurate delineations of anatomically relevant structures
are required for cancer treatment planning. Despite its
accuracy, manual labeling is time-consuming and
tedious-hence, the potential of automatic approaches, such
as deep learning models, is being investigated. A promising
trend in deep learning tumor segmentation is cross-modal
domain adaptation, where knowledge learned on one source
distribution (e.g., one modality) is transferred to another
distribution. Yet, artificial intelligence (AI) engineers
developing such models, need to thoroughly assess the
robustness of their approaches, which demands a deep
understanding of the model(s) behavior. In this paper, we
propose a web-based visual analytics application that
supports the visual assessment of the predictive performance
of deep learning-based models built for cross-modal brain
tumor segmentation. Our application supports the multi-level
comparison of multiple models drilling from entire cohorts
of patients down to individual slices, facilitates the
analysis of the relationship between image-derived features
and model performance, and enables the comparative
exploration of the predictive outcomes of the models. All
this is realized in an interactive interface with multiple
linked views. We present three use cases, analyzing
differences in deep learning segmentation approaches, the
influence of the tumor size, and the relationship of other
data set characteristics to the performance. From these
scenarios, we discovered that the tumor size, i.e., both
volumetric in 3D data and pixel count in 2D data, highly
affects the model performance, as samples with small tumors
often yield poorer results. Our approach is able to reveal
the best algorithms and their optimal configurations to
support AI engineers in obtaining more insights for the
development of their segmentation models.",
month = sep,
isbn = "978-3-03868-177-9",
publisher = "The Eurographics Association",
location = "Wien",
event = "Eurographics Workshop on Visual Computing for Biology and
Medicine (VCBM2022)",
doi = "10.2312/vcbm.20221193",
booktitle = "Eurographics Workshop on Visual Computing for Biology and
Medicine (VCBM 2022)",
pages = "5",
pages = "111--115",
keywords = "Visual Analytics, Life and medical sciences, Applied
computing",
URL = "https://www.cg.tuwien.ac.at/research/publications/2022/magg2022/",
}