Information
- Publication Type: Journal Paper (without talk)
- Workgroup(s)/Project(s):
- Date: June 2022
- DOI: 10.1016/j.cag.2022.04.014
- ISSN: 1873-7684
- Journal: Computers & Graphics
- Open Access: yes
- Pages: 13
- Volume: 105
- Publisher: Elsevier
- Pages: 12 – 24
- Keywords: networks, neuroscience, graph layouting, brain parcellation, anatomical layouts
Abstract
Recent advances in neuro-imaging enable scientists to create brain network data that can lead to novel insights into neurocircuitry, and a better understanding of the brain’s organization. These networks inherently involve a spatial component, depicting which brain regions are structurally, functionally or genetically related. Their visualization in 3D suffers from occlusion and clutter, especially with increasing number of nodes and connections, while 2D representations such as connectograms, connectivity matrices, and node-link diagrams neglect the spatio-anatomical context. Approaches to arrange 2D-graphs manually are tedious, species-dependent, and require the knowledge of domain experts.
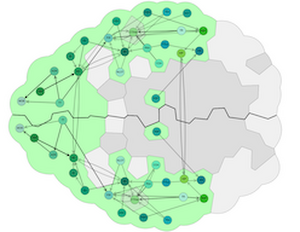
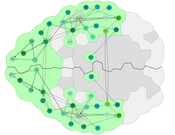
In this paper, we present a spatial-data-driven approach for layouting 3D brain networks in 2D node-link diagrams, while maintaining their spatial organization. The produced graphs do not need manual positioning of nodes, are consistent (even for sub-graphs), and provide a perspective-dependent arrangement for orientation. Furthermore, we provide a visual design for highlighting anatomical context, including the shape of the brain, and the size of brain regions. We present in several case-studies the applicability of our approach for different neuroscience-relevant species, including the mouse, human, and Drosophila larvae. In a user study conducted with several domain experts, we demonstrate its relevance and validity, as well as its potential for neuroscientific publications, presentations, and education.
Additional Files and Images
Additional images and videos
Additional files
Weblinks
BibTeX
@article{Ganglberger-2022-cg,
title = "Spatial-Data-Driven Layouting for Brain Network
Visualization",
author = "Florian Ganglberger and Monika Wi{\ss}mann and Hsiang-Yun Wu
and Nicolas Swoboda and Andreas Thum and Wulf Haubensak and
Katja B\"{u}hler",
year = "2022",
abstract = "Recent advances in neuro-imaging enable scientists to create
brain network data that can lead to novel insights into
neurocircuitry, and a better understanding of the brain’s
organization. These networks inherently involve a spatial
component, depicting which brain regions are structurally,
functionally or genetically related. Their visualization in
3D suffers from occlusion and clutter, especially with
increasing number of nodes and connections, while 2D
representations such as connectograms, connectivity
matrices, and node-link diagrams neglect the
spatio-anatomical context. Approaches to arrange 2D-graphs
manually are tedious, species-dependent, and require the
knowledge of domain experts. In this paper, we present a
spatial-data-driven approach for layouting 3D brain networks
in 2D node-link diagrams, while maintaining their spatial
organization. The produced graphs do not need manual
positioning of nodes, are consistent (even for sub-graphs),
and provide a perspective-dependent arrangement for
orientation. Furthermore, we provide a visual design for
highlighting anatomical context, including the shape of the
brain, and the size of brain regions. We present in several
case-studies the applicability of our approach for different
neuroscience-relevant species, including the mouse, human,
and Drosophila larvae. In a user study conducted with
several domain experts, we demonstrate its relevance and
validity, as well as its potential for neuroscientific
publications, presentations, and education.",
month = jun,
doi = "10.1016/j.cag.2022.04.014",
issn = "1873-7684",
journal = "Computers & Graphics",
pages = "13",
volume = "105",
publisher = "Elsevier",
pages = "12--24",
keywords = "networks, neuroscience, graph layouting, brain parcellation,
anatomical layouts",
URL = "https://www.cg.tuwien.ac.at/research/publications/2022/Ganglberger-2022-cg/",
}