Information
- Publication Type: WorkshopTalk
- Workgroup(s)/Project(s):
- Date: 20. October 2019
- Call for Papers: Call for Paper
- Event: Vis 2019 Workshop
- Lecturer: Hsiang-Yun Wu
- Location: Canada
- Keywords: Graph drawing, multilayer network, biological pathway
Abstract
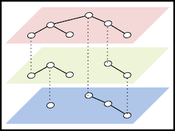
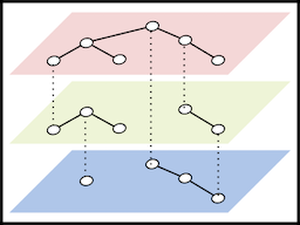
The concept of multilayer networks has become recently integrated into complex systems modeling since it encapsulates a very general concept of complex relationships. Biological pathways are an exam- ple of complex real-world networks, where vertices represent biolog- ical entities, and edges indicate the underlying connectivity. For this reason, using multilayer networks to model biological knowledge allows us to formally cover essential properties and theories in the field, which also raises challenges in visualization. This is because, in the early days of pathway visualization research, only restricted types of graphs, such as simple graphs, clustered graphs, and others were adopted. In this paper, we revisit a heterogeneous definition of biological networks and aim to provide an overview to see the gaps between data modeling and visual representation. The contribution will, therefore, lie in providing guidelines and challenges of using multilayer networks as a unified data structure for the biological pathway visualization.Additional Files and Images
Weblinks
No further information available.BibTeX
@WorkshopTalk{wu-2019-visworkshop,
title = "Graph Models for Biological Pathway Visualization: State of
the Art and Future Challenges",
author = "Hsiang-Yun Wu and Martin N\"{o}llenburg and Ivan Viola",
year = "2019",
abstract = "The concept of multilayer networks has become recently
integrated into complex systems modeling since it
encapsulates a very general concept of complex
relationships. Biological pathways are an exam- ple of
complex real-world networks, where vertices represent
biolog- ical entities, and edges indicate the underlying
connectivity. For this reason, using multilayer networks to
model biological knowledge allows us to formally cover
essential properties and theories in the field, which also
raises challenges in visualization. This is because, in the
early days of pathway visualization research, only
restricted types of graphs, such as simple graphs, clustered
graphs, and others were adopted. In this paper, we revisit a
heterogeneous definition of biological networks and aim to
provide an overview to see the gaps between data modeling
and visual representation. The contribution will, therefore,
lie in providing guidelines and challenges of using
multilayer networks as a unified data structure for the
biological pathway visualization. ",
month = oct,
event = "Vis 2019 Workshop",
location = "Canada",
keywords = "Graph drawing, multilayer network, biological pathway",
URL = "https://www.cg.tuwien.ac.at/research/publications/2019/wu-2019-visworkshop/",
}

 paper
paper