Information
- Publication Type: Journal Paper (without talk)
- Workgroup(s)/Project(s):
- Date: May 2019
- DOI: http://doi.org/10.1186/s12859-019-2779-4
- Journal: BMC Bioinformatics
- Number: 187
- Volume: 20
- Pages: 1 – 20
- Keywords: Biological pathways, Graph drawing, Mapmetaphor, Orthogonallayout, Floorplanning, Edgerouting
Abstract
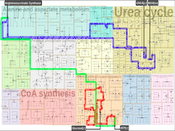
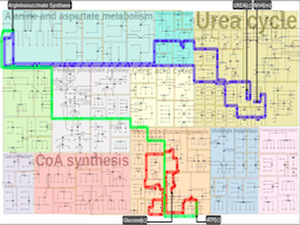
Background Biological pathways represent chains of molecular interactions in biological systems that jointly form complex dynamic networks. The network structure changes from the significance of biological experiments and layout algorithms often sacrifice low-level details to maintain high-level information, which complicates the entire image to large biochemical systems such as human metabolic pathways.Results Our work is inspired by concepts from urban planning since we create a visual hierarchy of biological pathways, which is analogous to city blocks and grid-like road networks in an urban area. We automatize the manual drawing process of biologists by first partitioning the map domain into multiple sub-blocks, and then building the corresponding pathways by routing edges schematically, to maintain the global and local context simultaneously. Our system incorporates constrained floor-planning and network-flow algorithms to optimize the layout of sub-blocks and to distribute the edge density along the map domain. We have developed the approach in close collaboration with domain experts and present their feedback on the pathway diagrams based on selected use cases.
Conclusions We present a new approach for computing biological pathway maps that untangles visual clutter by decomposing large networks into semantic sub-networks and bundling long edges to create space for presenting relationships systematically.
Additional Files and Images
Weblinks
BibTeX
@article{wu-2019-bmc,
title = "Metabopolis: Scalable Network Layout for Biological Pathway
Diagrams in Urban Map Style",
author = "Hsiang-Yun Wu and Martin N\"{o}llenburg and Filipa L. Sousa
and Ivan Viola",
year = "2019",
abstract = "Background Biological pathways represent chains of molecular
interactions in biological systems that jointly form complex
dynamic networks. The network structure changes from the
significance of biological experiments and layout algorithms
often sacrifice low-level details to maintain high-level
information, which complicates the entire image to large
biochemical systems such as human metabolic pathways.
Results Our work is inspired by concepts from urban planning
since we create a visual hierarchy of biological pathways,
which is analogous to city blocks and grid-like road
networks in an urban area. We automatize the manual drawing
process of biologists by first partitioning the map domain
into multiple sub-blocks, and then building the
corresponding pathways by routing edges schematically, to
maintain the global and local context simultaneously. Our
system incorporates constrained floor-planning and
network-flow algorithms to optimize the layout of sub-blocks
and to distribute the edge density along the map domain. We
have developed the approach in close collaboration with
domain experts and present their feedback on the pathway
diagrams based on selected use cases. Conclusions We
present a new approach for computing biological pathway maps
that untangles visual clutter by decomposing large networks
into semantic sub-networks and bundling long edges to create
space for presenting relationships systematically.",
month = may,
doi = "http://doi.org/10.1186/s12859-019-2779-4",
journal = "BMC Bioinformatics",
number = "187",
volume = "20",
pages = "1--20",
keywords = "Biological pathways, Graph drawing, Mapmetaphor,
Orthogonallayout, Floorplanning, Edgerouting",
URL = "https://www.cg.tuwien.ac.at/research/publications/2019/wu-2019-bmc/",
}

 paper
paper