Information
- Publication Type: Student Project
- Workgroup(s)/Project(s):
- Date: 2019
- Date (Start): October 2018
- Date (End): February 2019
- Matrikelnummer: 01325652
- First Supervisor: Manuela Waldner
- Keywords: pathways, web-based visualization
Abstract
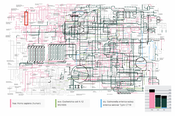
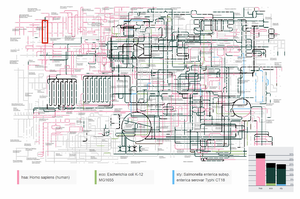
The Kyoto Encyclopaedia of Genes and Genomes (KEGG) resource is a combination of multiple databases, containing information about biochemical compounds, reactions, pathways, genes and much more. This database is one of the main resources for bioinformaticians and biologists to gain an understanding of molecular functionality inside organisms. The Orthology (KO) database from KEGG assigns pathways and genes with identical functionality to the same ortholog groups (KO entries). Therefore it is possible to map genes onto the pathway maps and obtain organism-specific visualizations. KEGG offers a web-based graph visualization to explore these pathways, however, the interaction possibilities are restricted and the rendering is inefficient. It is possible to visualize organism-specific pathways but a visual analysis tool to compare ortholog groups of multiple organisms is missing. In this work, we present an efficient interactive web application to compare ortholog groups of multiple organisms in the metabolic reference pathway. We introduce a graph overlay technique to mark the differences and similarities between multiple organisms and demonstrate it with two use cases. Additionally, we compare it against an existing point set membership visualization.Additional Files and Images
Additional images and videos
Additional files
Weblinks
- Online tool
Best to be used with Chrome. Enter the organisms to be compared by entering them as a string like this: "eco+hsa+mmu" (compares E-Coli, Homo Sapiens, and Mus Musculus). - List of KEGG organisms
List of organisms available in KEGG. - CESCG paper
Paper presented at the Central European Seminar on Computer Graphics 2019.
BibTeX
@studentproject{unger-2019_vcp,
title = "Visual Comparison of Organism-Specific Metabolic Pathways",
author = "Katharina Unger",
year = "2019",
abstract = "The Kyoto Encyclopaedia of Genes and Genomes (KEGG) resource
is a combination of multiple databases, containing
information about biochemical compounds, reactions,
pathways, genes and much more. This database is one of the
main resources for bioinformaticians and biologists to gain
an understanding of molecular functionality inside
organisms. The Orthology (KO) database from KEGG assigns
pathways and genes with identical functionality to the same
ortholog groups (KO entries). Therefore it is possible to
map genes onto the pathway maps and obtain organism-specific
visualizations. KEGG offers a web-based graph visualization
to explore these pathways, however, the interaction
possibilities are restricted and the rendering is
inefficient. It is possible to visualize organism-specific
pathways but a visual analysis tool to compare ortholog
groups of multiple organisms is missing. In this work, we
present an efficient interactive web application to compare
ortholog groups of multiple organisms in the metabolic
reference pathway. We introduce a graph overlay technique to
mark the differences and similarities between multiple
organisms and demonstrate it with two use cases.
Additionally, we compare it against an existing point set
membership visualization.",
month = feb,
keywords = "pathways, web-based visualization",
URL = "https://www.cg.tuwien.ac.at/research/publications/2019/unger-2019_vcp/",
}
 CESCG paper
CESCG paper