Information
- Publication Type: PhD-Thesis
- Workgroup(s)/Project(s):
- Date: November 2019
- Date (Start): 2016
- Date (End): 2019
- TU Wien Library:
- Second Supervisor: Eduard Gröller

- Open Access: yes
- 1st Reviewer: Thomas Ertl
- 2nd Reviewer: Timo Ropinski
- Rigorosum: 29. November 2019
- First Supervisor: Ivan Viola

Abstract
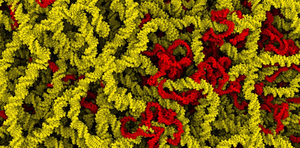
AbstractComputational models have advanced research of integrative cell biology in variousways. Especially in the biological mesoscale, the scale between atoms and cellularenvironments, computational models improve the understanding and qualitative anal-ysis. The mesoscale is an important range, since it represents the range of scalesthat are not fully accessible to a single experimental technique. Complex molecularassemblies within this scale have been visualized with x-ray crystallography, thoughonly in isolation. Mesoscale models shows how molecules are assembled into morecomplex subcelluar environments that orchestrate the processes of life. The skillfulcombination of the results of imaging and experimental techniques provides a glimpseof the processes, which are happening here. Only recently, biologists have startedto unify the various sources of information. They have begun to computationallyassemble and subsequently visualize complex environments, such as viruses or bacteria.Currently, we live in an opportune time for researching integrative structural biologydue to several factors. First and foremost, the wealth of data, driven through sourceslike online databases, makes structural information about biological entities publiclyavailable. In addition to that, the progress of parallel processors builds the foundationto instantly construct and render large mesoscale environments in atomistic detail.Finally, new scientific advances in visualization allow the efficient rendering of complexbiological phenomena with millions of structural units.In this cumulative thesis, we propose several novel techniques that facilitate the instantconstruction of mesoscale structures. The common methodological strategy of thesetechniques and insight from this thesis is “compute instead of store”. This approacheliminates the storage and memory management complexity, and enables instantchanges of the constructed models. Combined, our techniques are capable of instantlyconstructing large-scale biological environments using the basic structural buildingblocks of cells. These building blocks are mainly nucleic acids, lipids, and solubleproteins. For the generation of long linear polymers formed by nucleic acids, wepropose a parallel construction technique that makes use of a midpoint displacementalgorithm. The efficient generation of lipid membranes is realized through a texturesynthesis approach that makes use of the Wang tiling concept. For the population ofsoluble proteins, we present a staged algorithm, whereby each stage is processed inparallel. We have integrated the instant construction approach into a visual environmentin order to improve several aspects. First, it allows immediate feedback on the createdix
structures and the results of parameter changes. Additionally, the integration ofconstruction in visualization builds the foundation for visualization systems that striveto construct large-scale environments on-the-fly. Lastly, it advances the qualitativeanalysis of biological mesoscale environments, where a multitude of synthesized modelsis required. In order to disseminate the physiology of biological mesoscale models,we propose a novel concept that simplifies the creation of multi-scale proceduralanimations.
Additional Files and Images
Additional images and videos
Additional files
Weblinks
BibTeX
@phdthesis{klein_2019_PHD,
title = "Instant Construction of Atomistic Models for Visualization
in Integrative Cell Biology",
author = "Tobias Klein",
year = "2019",
abstract = "AbstractComputational models have advanced research of
integrative cell biology in variousways. Especially in the
biological mesoscale, the scale between atoms and
cellularenvironments, computational models improve the
understanding and qualitative anal-ysis. The mesoscale
is an important range, since it represents the range
of scalesthat are not fully accessible to a single
experimental technique. Complex molecularassemblies within
this scale have been visualized with x-ray crystallography,
thoughonly in isolation. Mesoscale models shows how
molecules are assembled into morecomplex subcelluar
environments that orchestrate the processes of life. The
skillfulcombination of the results of imaging and
experimental techniques provides a glimpseof the processes,
which are happening here. Only recently, biologists have
startedto unify the various sources of information.
They have begun to computationallyassemble and
subsequently visualize complex environments, such as viruses
or bacteria.Currently, we live in an opportune time for
researching integrative structural biologydue to several
factors. First and foremost, the wealth of data, driven
through sourceslike online databases, makes structural
information about biological entities publiclyavailable. In
addition to that, the progress of parallel processors builds
the foundationto instantly construct and render large
mesoscale environments in atomistic detail.Finally, new
scientific advances in visualization allow the efficient
rendering of complexbiological phenomena with millions of
structural units.In this cumulative thesis, we propose
several novel techniques that facilitate the
instantconstruction of mesoscale structures. The common
methodological strategy of thesetechniques and insight from
this thesis is “compute instead of store”. This
approacheliminates the storage and memory management
complexity, and enables instantchanges of the constructed
models. Combined, our techniques are capable of
instantlyconstructing large-scale biological environments
using the basic structural buildingblocks of cells. These
building blocks are mainly nucleic acids, lipids, and
solubleproteins. For the generation of long linear
polymers formed by nucleic acids, wepropose a parallel
construction technique that makes use of a midpoint
displacementalgorithm. The efficient generation of lipid
membranes is realized through a texturesynthesis approach
that makes use of the Wang tiling concept. For the
population ofsoluble proteins, we present a staged
algorithm, whereby each stage is processed inparallel. We
have integrated the instant construction approach into a
visual environmentin order to improve several aspects.
First, it allows immediate feedback on the createdix
structures and the results of parameter changes.
Additionally, the integration ofconstruction in
visualization builds the foundation for visualization
systems that striveto construct large-scale environments
on-the-fly. Lastly, it advances the qualitativeanalysis of
biological mesoscale environments, where a multitude of
synthesized modelsis required. In order to disseminate the
physiology of biological mesoscale models,we propose a
novel concept that simplifies the creation of
multi-scale proceduralanimations. ",
month = nov,
address = "Favoritenstrasse 9-11/E193-02, A-1040 Vienna, Austria",
school = "Research Unit of Computer Graphics, Institute of Visual
Computing and Human-Centered Technology, Faculty of
Informatics, TU Wien ",
URL = "https://www.cg.tuwien.ac.at/research/publications/2019/klein_2019_PHD/",
}

 image
image
