Information
- Publication Type: Bachelor Thesis
- Workgroup(s)/Project(s):
- Date: June 2017
- Date (Start): November 2017
- Date (End): June 2017
- Matrikelnummer: 01635324
- First Supervisor:
Abstract
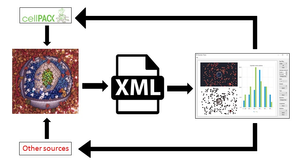
Currently available techniques for capturing macromolecules on atomic level are not appropriate for large structures on the biological mesoscale. Therefore, those structures, such as viruses or cell organelles, have to be assembled from molecular building blocks using software tools. The goal of recent projects like cellPACK is to create models with these tools, allowing the scientific community to iteratively give feedback and edit the models, in order to eventually generate the most suitable illustration consistent with the current state of knowledge. For that purpose, we need to discern the values for properties like distribution, density or opacity that make a model preferable to others. This thesis aims to create a software program for visual evaluation of the quality of the assembled structures. The program will extract the information about the quality of spatial distribution of molecules in the scenes produced by packing tools and plot it into a set of 2D representations. These will convey the statistical information about the distribution and enable the visual comparison of generated models, which vary not only due to the stochastic nature of the packing algorithms but also because of the use of different parameter settings.Additional Files and Images
Weblinks
No further information available.BibTeX
@bachelorsthesis{Escribano_2017,
title = "Visual Evaluation of Computational Models of the Biological
Mesoscale",
author = "Guillermo Garcia-Escribano",
year = "2017",
abstract = "Currently available techniques for capturing macromolecules
on atomic level are not appropriate for large structures on
the biological mesoscale. Therefore, those structures, such
as viruses or cell organelles, have to be assembled from
molecular building blocks using software tools. The goal of
recent projects like cellPACK is to create models with these
tools, allowing the scientific community to iteratively give
feedback and edit the models, in order to eventually
generate the most suitable illustration consistent with the
current state of knowledge. For that purpose, we need to
discern the values for properties like distribution, density
or opacity that make a model preferable to others. This
thesis aims to create a software program for visual
evaluation of the quality of the assembled structures. The
program will extract the information about the quality of
spatial distribution of molecules in the scenes produced by
packing tools and plot it into a set of 2D representations.
These will convey the statistical information about the
distribution and enable the visual comparison of generated
models, which vary not only due to the stochastic nature of
the packing algorithms but also because of the use of
different parameter settings.",
month = jun,
address = "Favoritenstrasse 9-11/E193-02, A-1040 Vienna, Austria",
school = "Institute of Computer Graphics and Algorithms, Vienna
University of Technology ",
URL = "https://www.cg.tuwien.ac.at/research/publications/2017/Escribano_2017/",
}