Information
- Publication Type: PhD-Thesis
- Workgroup(s)/Project(s):
- Date: 2009
- Date (Start): March 2006
- Date (End): January 2009
- TU Wien Library:
- Second Supervisor: Ivan Viola
- Rigorosum: 27. January 2009
- First Supervisor: Eduard Gröller
Abstract
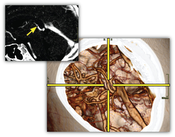
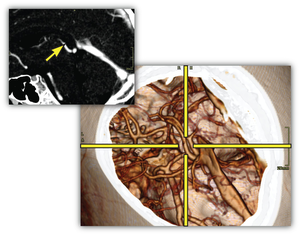
In this thesis two techniques for the smart linking of 2D and 3D views in medical applications are presented. Although real-time interactive 3D volume visualization is available even for very large data sets, it is used quite rarely in the clinical practice. A major obstacle for a better integration in the clinical workflow is the time-consuming process to adjust the parameters to generate diagnostically relevant images. The clinician has to take care of the appropriate viewpoint, zooming, transfer function setup, clipping planes, and other parameters. Because of this, current applications primarily employ 2D views generated through standard techniques such as multi-planar reformatting (MPR).The LiveSync interaction metaphor is a new concept to synchronize 2D slice views and 3D volumetric views of medical data sets. Through intuitive picking actions on the slice, the users define the anatomical structures they are interested in. The 3D volumetric view is updated automatically with the goal that the users are provided with diagnostically relevant images. To achieve this live synchronization a minimal set of derived information, without the need for segmented data sets or data-specific precomputations, is used. The presented system provides the physician with synchronized views which help to gain deeper insight into the medical data with minimal user interaction.
Contextual picking is a novel method for the interactive identification of contextual interest points within volumetric data by picking on a direct volume rendered image. In clinical diagnostics the points of interest are often located in the center of anatomical structures. In order to derive the volumetric position, which allows a convenient examination of the intended structure, the system automatically extracts contextual meta information from the DICOM (Digital Imaging and Communications in Medicine) images and the setup of the medical workstation. Along a viewing ray for a volumetric picking, the ray profile is analyzed to detect structures which are similar to predefined templates from a knowledge base. It is demonstrated that the obtained position in 3D can be utilized to highlight a structure in 2D slice views, to interactively calculate approximate centerlines of tubular objects, or to place labels at contextually-defined 3D positions.
Additional Files and Images
Weblinks
No further information available.BibTeX
@phdthesis{kohlmann-2009-lssl,
title = "LiveSync: Smart Linking of 2D and 3D Views in Medical
Applications",
author = "Peter Kohlmann",
year = "2009",
abstract = "In this thesis two techniques for the smart linking of 2D
and 3D views in medical applications are presented. Although
real-time interactive 3D volume visualization is available
even for very large data sets, it is used quite rarely in
the clinical practice. A major obstacle for a better
integration in the clinical workflow is the time-consuming
process to adjust the parameters to generate diagnostically
relevant images. The clinician has to take care of the
appropriate viewpoint, zooming, transfer function setup,
clipping planes, and other parameters. Because of this,
current applications primarily employ 2D views generated
through standard techniques such as multi-planar
reformatting (MPR). The LiveSync interaction metaphor is a
new concept to synchronize 2D slice views and 3D volumetric
views of medical data sets. Through intuitive picking
actions on the slice, the users define the anatomical
structures they are interested in. The 3D volumetric view is
updated automatically with the goal that the users are
provided with diagnostically relevant images. To achieve
this live synchronization a minimal set of derived
information, without the need for segmented data sets or
data-specific precomputations, is used. The presented system
provides the physician with synchronized views which help to
gain deeper insight into the medical data with minimal user
interaction. Contextual picking is a novel method for the
interactive identification of contextual interest points
within volumetric data by picking on a direct volume
rendered image. In clinical diagnostics the points of
interest are often located in the center of anatomical
structures. In order to derive the volumetric position,
which allows a convenient examination of the intended
structure, the system automatically extracts contextual meta
information from the DICOM (Digital Imaging and
Communications in Medicine) images and the setup of the
medical workstation. Along a viewing ray for a volumetric
picking, the ray profile is analyzed to detect structures
which are similar to predefined templates from a knowledge
base. It is demonstrated that the obtained position in 3D
can be utilized to highlight a structure in 2D slice views,
to interactively calculate approximate centerlines of
tubular objects, or to place labels at contextually-defined
3D positions.",
address = "Favoritenstrasse 9-11/E193-02, A-1040 Vienna, Austria",
school = "Institute of Computer Graphics and Algorithms, Vienna
University of Technology ",
URL = "https://www.cg.tuwien.ac.at/research/publications/2009/kohlmann-2009-lssl/",
}

 image
image screen version
screen version