| (25) |
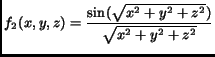 |
(26) |
| (27) |
In order to evaluate the quality of the gradient approximation we used three different analytical functions for comparison purposes:
| (25) |
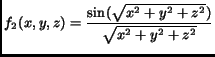 |
(26) |
| (27) |
The results of our experiment can be seen in Fig 7 (a) - (c). The first row shows the relative error in magnitude and the second row shows the angular error. Column one depicts the error of our first gradient reconstruction method (Eq. 22) that is based on central differences at the grid point itself. Column 2 corresponds to method two (Eq. 23), which is the average of all central differences at the of the cube edges surrounding the sampling point. In the last column we computed the linearly interpolated central differences, assuming the data set was given on a regular grid with corresponding dimensions.
Fig 7(a) shows the error images for function ![]() . In
this image an angular error of 15 degrees and an amplitude error of
30% corresponds to white (255). Fig. 7(b) shows the
error images for function
. In
this image an angular error of 15 degrees and an amplitude error of
30% corresponds to white (255). Fig. 7(b) shows the
error images for function ![]() . Here an angular error of 30 degrees
and an amplitude error of 60% corresponds to white (255). Finally the
results for function
. Here an angular error of 30 degrees
and an amplitude error of 60% corresponds to white (255). Finally the
results for function ![]() are displayed in Fig. 7(c).
Here 5 degrees for the angular error and 10% for the amplitude error
correspond to white (255).
are displayed in Fig. 7(c).
Here 5 degrees for the angular error and 10% for the amplitude error
correspond to white (255).
![\includegraphics[width=1.0\columnwidth]{pics/sphere57.eps}](img77.gif)
(a) ![\includegraphics[width=1.0\columnwidth]{pics/sinc57.eps}](img78.gif)
(b) ![\includegraphics[width=1.0\columnwidth]{pics/ml57.eps}](img79.gif)
(c) |
From these images we conclude that both our difference methods are quite comparable with central differencing and linear interpolation on regular grids. Hence one need not to worry about quality loss by using bcc grids for volume rendering applications. Furthermore since there are no large differences between the two introduced methods in Section 3.3, we don't find the expensive operations of method 2 justified.
Fig. 8 shows images of the Marschner-Lobb data set sampled
on a
![]() Cartesian grid (as described by Marschner
and Lobb [11]) on the left respectively a
Cartesian grid (as described by Marschner
and Lobb [11]) on the left respectively a
![]() bcc grid on the right. This data set is quite demanding
for a straightforward splatter and there are some visible differences
in the results. The image generated from the bcc grid is rather
blurred whereas the image from the Cartesian grid exhibits strong
artifacts, especially in diagonal directions.
bcc grid on the right. This data set is quite demanding
for a straightforward splatter and there are some visible differences
in the results. The image generated from the bcc grid is rather
blurred whereas the image from the Cartesian grid exhibits strong
artifacts, especially in diagonal directions.
The data sets that we used for rendering the images in Color
Plate 1 were produced using a high-quality
interpolation filter. We used the ![]() -4EF filter as designed by
Möller et al. [13]. In Color Plate 1 we
show results of rendering the ``neghip'' data set as well as the High
Potential Iron Protein data set by Louis Noodleman and David Case, of
Scripps Clinic, La Jolla, California, as well as the fuel injection
data set. Again, a regular Cartesian grid was used on the left and a
bcc grid on the right. There are some visible differences in the
images. Since we classify different values that represent two
different grid positions one cannot expect identical pictures. Hence
we see some differences resulting from the problem of
pre-classification [20].
-4EF filter as designed by
Möller et al. [13]. In Color Plate 1 we
show results of rendering the ``neghip'' data set as well as the High
Potential Iron Protein data set by Louis Noodleman and David Case, of
Scripps Clinic, La Jolla, California, as well as the fuel injection
data set. Again, a regular Cartesian grid was used on the left and a
bcc grid on the right. There are some visible differences in the
images. Since we classify different values that represent two
different grid positions one cannot expect identical pictures. Hence
we see some differences resulting from the problem of
pre-classification [20].
We also did some timings which are reported in Table 1. It is interesting to note that the speedup for some data sets were bigger than expected. This could have been caused by the decreased memory caching necessary. For a very small data set (lobster) we saw expected speedups near 30%.
Our results indicate that the resampled data have the potential to
lead to better compression. We were able to show that our compression
ratios for practical data sets are better than what was achieved using
the gzip utility. Also, our overall compression ratios were better
then previously reported [5]. Table 2
shows compression ratios of various volume data sets. Note that the
last two columns give percentages as compared to the original data
size indicating the overall compression ratio, which is what we are
interested in. However, the compression of synthetic data sets is a
rather surprising result and needs to be further investigated.