Information
- Publication Type: Journal Paper with Conference Talk
- Workgroup(s)/Project(s): not specified
- Date: October 2022
- Journal: IEEE Transactions on Visualization and Computer Graphics
- Lecturer: Pavol Ulbrich
- Event: IEEE VIS 2022
- DOI: 10.1109/TVCG.2022.3209411
- Pages: 10
- Publisher: Institute of Electrical and Electronics Engineers (IEEE)
- Conference date: October 2022
- Pages: 1 – 10
- Keywords: Molecular Dynamics, structure, node-based visualization, progressive analytics, proteins, Analytical models, Biological system modeling, Three-dimensional displays, Computational modeling, Task analysis, Animation
Abstract
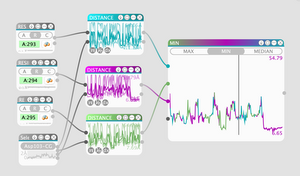
We present sMolBoxes, a dataflow representation for the exploration and analysis of long molecular dynamics (MD) simulations. When MD simulations reach millions of snapshots, a frame-by-frame observation is not feasible anymore. Thus, biochemists rely to a large extent only on quantitative analysis of geometric and physico-chemical properties. However, the usage of abstract methods to study inherently spatial data hinders the exploration and poses a considerable workload. sMolBoxes link quantitative analysis of a user-defined set of properties with interactive 3D visualizations. They enable visual explanations of molecular behaviors, which lead to an efficient discovery of biochemically significant parts of the MD simulation. sMolBoxes follow a node-based model for flexible definition, combination, and immediate evaluation of properties to be investigated. Progressive analytics enable fluid switching between multiple properties, which facilitates hypothesis generation. Each sMolBox provides quick insight to an observed property or function, available in more detail in the bigBox View. The case studies illustrate that even with relatively few sMolBoxes, it is possible to express complex analytical tasks, and their use in exploratory analysis is perceived as more efficient than traditional scripting-based methods.
Additional Files and Images
Additional images and videos
Additional files
Weblinks
BibTeX
@article{sMolBoxes_2022,
title = "sMolBoxes: Dataflow Model for Molecular Dynamics Exploration",
author = "Pavol Ulbrich and Manuela Waldner and Katar\'{i}na
Furmanov\'{a} and Sergio M. Margues and David Bedn\'{a}\v{r}
and Barbora Kozlikova and Jan Byska",
year = "2022",
abstract = "We present sMolBoxes, a dataflow representation for the
exploration and analysis of long molecular dynamics (MD)
simulations. When MD simulations reach millions of
snapshots, a frame-by-frame observation is not feasible
anymore. Thus, biochemists rely to a large extent only on
quantitative analysis of geometric and physico-chemical
properties. However, the usage of abstract methods to study
inherently spatial data hinders the exploration and poses a
considerable workload. sMolBoxes link quantitative analysis
of a user-defined set of properties with interactive 3D
visualizations. They enable visual explanations of molecular
behaviors, which lead to an efficient discovery of
biochemically significant parts of the MD simulation.
sMolBoxes follow a node-based model for flexible definition,
combination, and immediate evaluation of properties to be
investigated. Progressive analytics enable fluid switching
between multiple properties, which facilitates hypothesis
generation. Each sMolBox provides quick insight to an
observed property or function, available in more detail in
the bigBox View. The case studies illustrate that even with
relatively few sMolBoxes, it is possible to express complex
analytical tasks, and their use in exploratory analysis is
perceived as more efficient than traditional scripting-based
methods.",
month = oct,
journal = "IEEE Transactions on Visualization and Computer Graphics",
doi = "10.1109/TVCG.2022.3209411",
pages = "10",
publisher = "Institute of Electrical and Electronics Engineers (IEEE)",
pages = "1--10",
keywords = "Molecular Dynamics, structure, node-based visualization,
progressive analytics, proteins, Analytical models,
Biological system modeling, Three-dimensional displays,
Computational modeling, Task analysis, Animation",
URL = "https://www.cg.tuwien.ac.at/research/publications/2022/sMolBoxes_2022/",
}