Information
- Publication Type: PhD-Thesis
- Workgroup(s)/Project(s):
- Date: April 2021
- Date (Start): March 2017
- Date (End): April 2021
- Open Access: yes
- 1st Reviewer: Kwan Liu Ma
- 2nd Reviewer: Bernhard Preim
- Rigorosum: 7. April 2021
- First Supervisor: Ivan Viola

- Pages: 123
- Keywords: biological, visualization, multi-scale, dense, navigation
Abstract
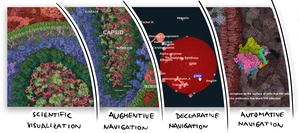
This thesis focuses on the problem of navigating complex 3D models composed of large numbers of molecular instances packed densely in the three-dimensional space. Due to the large environment encompassing several magnitudes of scale, the traditional navigational paradigms applied in real-time computer graphics are becoming insufficient when applied to biological environments. In the first part of the thesis, I analyze navigation challenges presented by such a use case and recognize several modes of navigation that can be employed when interfacing a complex 3D visualization system with the end-user. We discuss three such modes of navigation: augmentive, declarative, and automative. First, in augmentive navigation, the user is given fully manual control over all aspects of navigation, such as controlling the camera's position and rotation, or the visibility of the individual model's parts. Their manual exploration is, however, augmented by automatically deployed annotation to help make sense of the environment. In declarative navigation, the user interaction is simplified to declaring their target. The visualization system then takes over the low-level controls of the visualization, e.g., camera path animation and scene visibility transition, which are computed to navigate the user directly to their declared target. The third stage, automative navigation, relieves the user from even this responsibility and places the choice of what gets shown to an algorithmic solution. In this case, such automated fly-through can then be guided by a specific storyline. In the second part of the thesis, I present specific methods addressing technical gaps and contributing to realizing the navigational stages presented in the first part. I start by introducing an approach for textual labeling of multi-scale molecular models inspired by the level-of-detail concept. That way, a scenario of augmentive navigation is provided. Second, I propose a navigational method for traversing a dense molecular model with a hierarchical organization, implementing the declarative navigation concept. The presented method uses textual labels for browsing the three-dimensional model, essentially providing a way of traversing the hierarchical organization and exploring the spatial characteristics of the model. Finally, I propose a pipeline for producing automated tours of molecular models, demonstrating the automative navigation mode.
Additional Files and Images
Additional images and videos
Additional files
Weblinks
BibTeX
@phdthesis{kouril-2021-phdthesis,
title = "Interactive Visualization of Dense and Multi-Scale Data for
Science Outreach",
author = "David Kou\v{r}il",
year = "2021",
abstract = "This thesis focuses on the problem of navigating complex 3D
models composed of large numbers of molecular instances
packed densely in the three-dimensional space. Due to the
large environment encompassing several magnitudes of scale,
the traditional navigational paradigms applied in real-time
computer graphics are becoming insufficient when applied to
biological environments. In the first part of the thesis, I
analyze navigation challenges presented by such a use case
and recognize several modes of navigation that can be
employed when interfacing a complex 3D visualization system
with the end-user. We discuss three such modes of
navigation: augmentive, declarative, and automative. First,
in augmentive navigation, the user is given fully manual
control over all aspects of navigation, such as controlling
the camera's position and rotation, or the visibility of the
individual model's parts. Their manual exploration is,
however, augmented by automatically deployed annotation to
help make sense of the environment. In declarative
navigation, the user interaction is simplified to declaring
their target. The visualization system then takes over the
low-level controls of the visualization, e.g., camera path
animation and scene visibility transition, which are
computed to navigate the user directly to their declared
target. The third stage, automative navigation, relieves the
user from even this responsibility and places the choice of
what gets shown to an algorithmic solution. In this case,
such automated fly-through can then be guided by a specific
storyline. In the second part of the thesis, I present
specific methods addressing technical gaps and contributing
to realizing the navigational stages presented in the first
part. I start by introducing an approach for textual
labeling of multi-scale molecular models inspired by the
level-of-detail concept. That way, a scenario of augmentive
navigation is provided. Second, I propose a navigational
method for traversing a dense molecular model with a
hierarchical organization, implementing the declarative
navigation concept. The presented method uses textual labels
for browsing the three-dimensional model, essentially
providing a way of traversing the hierarchical organization
and exploring the spatial characteristics of the model.
Finally, I propose a pipeline for producing automated tours
of molecular models, demonstrating the automative navigation
mode.",
month = apr,
pages = "123",
address = "Favoritenstrasse 9-11/E193-02, A-1040 Vienna, Austria",
school = "Research Unit of Computer Graphics, Institute of Visual
Computing and Human-Centered Technology, Faculty of
Informatics, TU Wien ",
keywords = "biological, visualization, multi-scale, dense, navigation",
URL = "https://www.cg.tuwien.ac.at/research/publications/2021/kouril-2021-phdthesis/",
}

 teaser
teaser thesis
thesis
