Information
- Publication Type: Journal Paper (without talk)
- Workgroup(s)/Project(s):
- Date: March 2021
- DOI: 10.1109/TVCG.2021.3065443
- Journal: IEEE TRANSACTIONS ON VISUALIZATION AND COMPUTER GRAPHICS
- Open Access: yes
- Volume: 3
- Pages: 1 – 13
Abstract
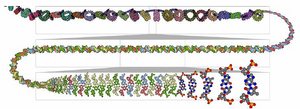
We present Multiscale Unfolding, an interactive technique for illustratively visualizing multiple hierarchical scales of DNA in a single view, showing the genome at different scales and demonstrating how one scale spatially folds into the next. The DNA’s extremely long sequential structure—arranged differently on several distinct scale levels—is often lost in traditional 3D depictions, mainly due to its multiple levels of dense spatial packing and the resulting occlusion. Furthermore, interactive exploration of this complex structure is cumbersome, requiring visibility management like cut-aways. In contrast to existing temporally controlled multiscale data exploration, we allow viewers to always see and interact with any of the involved scales. For this purpose we separate the depiction into constant-scale and scale transition zones. Constant-scale zones maintain a single-scale representation, while still linearly unfolding the DNA. Inspired by illustration, scale transition zones connect adjacent constant-scale zones via level unfolding, scaling, and transparency. We thus represent the spatial structure of the whole DNA macro-molecule, maintain its local organizational characteristics, linearize its higher-level organization, and use spatially controlled, understandable interpolation between neighboring scales. We also contribute interaction
techniques that provide viewers with a coarse-to-fine control for navigating within our all-scales-in-one-view representations and visual
aids to illustrate the size differences. Overall, Multiscale Unfolding allows viewers to grasp the DNA’s structural composition from
chromosomes to the atoms, with increasing levels of “unfoldedness,” and can be applied in data-driven illustration and communication.
Additional Files and Images
Additional images and videos
Additional files
Weblinks
BibTeX
@article{Sarkis_2021,
title = "Multiscale Unfolding: Illustratively Visualizing the Whole
Genome at a Glance",
author = "Sarkis Halladjian and David Kou\v{r}il and Haichao Miao and
Eduard Gr\"{o}ller and Ivan Viola and Tobias Isenberg",
year = "2021",
abstract = "We present Multiscale Unfolding, an interactive technique
for illustratively visualizing multiple hierarchical scales
of DNA in a single view, showing the genome at different
scales and demonstrating how one scale spatially folds into
the next. The DNA’s extremely long sequential
structure—arranged differently on several distinct scale
levels—is often lost in traditional 3D depictions, mainly
due to its multiple levels of dense spatial packing and the
resulting occlusion. Furthermore, interactive exploration of
this complex structure is cumbersome, requiring visibility
management like cut-aways. In contrast to existing
temporally controlled multiscale data exploration, we allow
viewers to always see and interact with any of the involved
scales. For this purpose we separate the depiction into
constant-scale and scale transition zones. Constant-scale
zones maintain a single-scale representation, while still
linearly unfolding the DNA. Inspired by illustration, scale
transition zones connect adjacent constant-scale zones via
level unfolding, scaling, and transparency. We thus
represent the spatial structure of the whole DNA
macro-molecule, maintain its local organizational
characteristics, linearize its higher-level organization,
and use spatially controlled, understandable interpolation
between neighboring scales. We also contribute interaction
techniques that provide viewers with a coarse-to-fine
control for navigating within our all-scales-in-one-view
representations and visual aids to illustrate the size
differences. Overall, Multiscale Unfolding allows viewers to
grasp the DNA’s structural composition from chromosomes
to the atoms, with increasing levels of “unfoldedness,”
and can be applied in data-driven illustration and
communication. ",
month = mar,
doi = "10.1109/TVCG.2021.3065443",
journal = "IEEE TRANSACTIONS ON VISUALIZATION AND COMPUTER GRAPHICS ",
volume = "3",
pages = "1--13",
URL = "https://www.cg.tuwien.ac.at/research/publications/2021/Sarkis_2021/",
}

 Image
Image Paper
Paper
