Information
- Publication Type: Journal Paper (without talk)
- Workgroup(s)/Project(s):
- Date: March 2021
- DOI: 10.1016/j.compbiomed.2021.104344
- Journal: Computers in Biology and Medicine
- Open Access: yes
- Volume: 133
- Pages: 1 – 14
Abstract
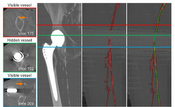
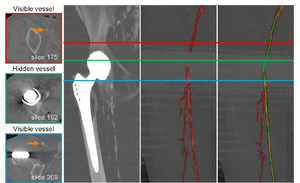
Objectives: Manual or semi-automated segmentation of the lower extremity arterial tree in patients with Pe-ripheral arterial disease (PAD) remains a notoriously difficult and time-consuming task. The complex manifes-tations of the disease, including discontinuities of the vascular flow channels, the presence of calcified atherosclerotic plaque in close vicinity to adjacent bone, and the presence of metal or other imaging artifacts currently preclude fully automated vessel identification. New machine learning techniques may alleviate this challenge, but require large and reasonably well segmented training data. Methods: We propose a novel semi-automatic vessel tracking approach for peripheral arteries to facilitate and accelerate the creation of annotated training data by expert cardiovascular radiologists or technologists, while limiting the number of necessary manual interactions, and reducing processing time. After automatically clas-sifying blood vessels, bones, and other tissue, the relevant vessels are tracked and organized in a tree-like structure for further visualization. Results: We conducted a pilot (N = 9) and a clinical study (N = 24) in which we assess the accuracy and required time for our approach to achieve sufficient quality for clinical application, with our current clinically established workflow as the standard of reference. Our approach enabled expert physicians to readily identify all clinically relevant lower extremity arteries, even in problematic cases, with an average sensitivity of 92.9%, and an average specificity and overall accuracy of 99.9%. Conclusions: Compared to the clinical workflow in our collaborating hospitals (28:40 ± 7:45 [mm:ss]), our approach (17:24 ± 6:44 [mm:ss]) is on average 11:16 [mm:ss] (39%) faster.Additional Files and Images
Weblinks
BibTeX
@article{Mistelbauer_2021,
title = "Semi-automatic vessel detection for challenging cases of
peripheral arterial disease ",
author = "Gabriel Mistelbauer and Anca Morar and R\"{u}diger
Schernthaner and Andreas Strassl and Dominik Fleischmann and
Florica Moldoveanu and Eduard Gr\"{o}ller",
year = "2021",
abstract = "Objectives: Manual or semi-automated segmentation of the
lower extremity arterial tree in patients with Pe-ripheral
arterial disease (PAD) remains a notoriously difficult and
time-consuming task. The complex manifes-tations of the
disease, including discontinuities of the vascular flow
channels, the presence of calcified atherosclerotic plaque
in close vicinity to adjacent bone, and the presence of
metal or other imaging artifacts currently preclude fully
automated vessel identification. New machine learning
techniques may alleviate this challenge, but require large
and reasonably well segmented training data. Methods: We
propose a novel semi-automatic vessel tracking approach for
peripheral arteries to facilitate and accelerate the
creation of annotated training data by expert cardiovascular
radiologists or technologists, while limiting the number of
necessary manual interactions, and reducing processing time.
After automatically clas-sifying blood vessels, bones, and
other tissue, the relevant vessels are tracked and organized
in a tree-like structure for further visualization.
Results: We conducted a pilot (N = 9) and a clinical study
(N = 24) in which we assess the accuracy and required time
for our approach to achieve sufficient quality for clinical
application, with our current clinically established
workflow as the standard of reference. Our approach enabled
expert physicians to readily identify all clinically
relevant lower extremity arteries, even in problematic
cases, with an average sensitivity of 92.9%, and an average
specificity and overall accuracy of 99.9%. Conclusions:
Compared to the clinical workflow in our collaborating
hospitals (28:40 ± 7:45 [mm:ss]), our approach (17:24 ±
6:44 [mm:ss]) is on average 11:16 [mm:ss] (39%) faster. ",
month = mar,
doi = "10.1016/j.compbiomed.2021.104344",
journal = "Computers in Biology and Medicine ",
volume = "133",
pages = "1--14",
URL = "https://www.cg.tuwien.ac.at/research/publications/2021/Mistelbauer_2021/",
}

 Image
Image Paper
Paper