Information
- Publication Type: Poster
- Workgroup(s)/Project(s):
- Date: June 2018
- Location: Rome
- Event: 3rd Functional DNA Nanotechnology Workshop
- Call for Papers: Call for Paper
- Conference date: 6. June 2018 – 8. June 2018
Abstract
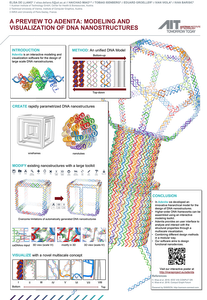
We present Adenita, an open-source software that aims to provide an integrated in silico design toolkit for DNA Nanostructures. It facilitates the modular assembly of pre-existing and de novo designs, regardless of the used approach. Adenita is being developed in the context of the MARA project [1], a highly ambitious project aiming to produce a DNA nanorobot capable of targeted cell lyses. Currently, the existing design and visualization tools are insufficient to solve the specific challenges in our project. We aim to overcome these limitations with Adenita, a new semi-automated approach that we are developing as the MARA project advances. Adenita integrates visualizations [2] with user interactions and algorithms in a semi-manual approach. At the core, we use a hierarchical data model that enables us to combine both a top-down (DAEDALUS [3]) and a bottom-up (caDNAno [4]) design approach of the DNA Nanostructure. From the DNA data model, we create smooth visualizations that depict the structure in multiple scales from its atomic details to a highlevel geometric representation of the target shape. In addition, we employ different layouts for the same structure [5]: 3D structural representations, 2D caDNAno-style diagrams, and 1D display of the linear sequences. Creators enable the parametrized generation of structural motifs, while Manipulators facilitate the advanced modifications of the structural properties, such as connecting components and adding bridging strands. Analysis of the designs is still in a preliminary phase, but it already enables the straightforward estimation of distances and the melting temperatures [6] of binding regions. The first DNA nanostructures that we designed with the new tool are now under experimental evaluation.Additional Files and Images
Weblinks
No further information available.BibTeX
@misc{Miao2018FDN,
title = "A Preview to Adenita: Visualization and Modeling of DNA
Nanostructures",
author = "Elisa De Llano and Haichao Miao and Tobias Isenberg and
Eduard Gr\"{o}ller and Ivan Viola and Ivan Barisic",
year = "2018",
abstract = "We present Adenita, an open-source software that aims to
provide an integrated in silico design toolkit for DNA
Nanostructures. It facilitates the modular assembly of
pre-existing and de novo designs, regardless of the used
approach. Adenita is being developed in the context of the
MARA project [1], a highly ambitious project aiming to
produce a DNA nanorobot capable of targeted cell lyses.
Currently, the existing design and visualization tools are
insufficient to solve the specific challenges in our
project. We aim to overcome these limitations with Adenita,
a new semi-automated approach that we are developing as the
MARA project advances. Adenita integrates visualizations [2]
with user interactions and algorithms in a semi-manual
approach. At the core, we use a hierarchical data model that
enables us to combine both a top-down (DAEDALUS [3]) and a
bottom-up (caDNAno [4]) design approach of the DNA
Nanostructure. From the DNA data model, we create smooth
visualizations that depict the structure in multiple scales
from its atomic details to a highlevel geometric
representation of the target shape. In addition, we employ
different layouts for the same structure [5]: 3D structural
representations, 2D caDNAno-style diagrams, and 1D display
of the linear sequences. Creators enable the parametrized
generation of structural motifs, while Manipulators
facilitate the advanced modifications of the structural
properties, such as connecting components and adding
bridging strands. Analysis of the designs is still in a
preliminary phase, but it already enables the
straightforward estimation of distances and the melting
temperatures [6] of binding regions. The first DNA
nanostructures that we designed with the new tool are now
under experimental evaluation. ",
month = jun,
location = "Rome",
event = "3rd Functional DNA Nanotechnology Workshop",
Conference date = "Poster presented at 3rd Functional DNA Nanotechnology
Workshop (2018-06-06--2018-06-08)",
URL = "https://www.cg.tuwien.ac.at/research/publications/2018/Miao2018FDN/",
}