Information
- Publication Type: Master Thesis
- Workgroup(s)/Project(s):
- Date: December 2017
- Date (Start): 2016
- Date (End): 28. November 2017
- TU Wien Library:
- First Supervisor:
Abstract
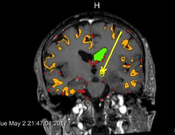
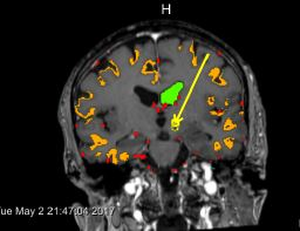
Neurosurgeons make decisions based on expert knowledge that takes factors such as safety margins, the avoidance of risk structures, trajectory length and trajectory angle into consideration. While some of those factors are mandatory, others can be optimized in order to obtain the best possible trajectory under the given circumstances. Through comparison with the actually chosen trajectories from real biopsies and qualitative interviews with domain experts, we identified important rules for trajectory planning. In this thesis, we present BrainXplore, an interactive visual analysis tool for aiding neurosurgeons in planning brain biopsies. BrainXplore is an extendable Biopsy Planning framework that incorporates those rules while at the same time leaving full flexibility for their customization and adding of new structures at risk. Automatically computed candidate trajectories can be incrementally refined in an interactive manner until an optimal trajectory is found. We employ a spatial index server as part of our system that allows us to access distance information on an unlimited number of risk structures at arbitrary resolution. Furthermore, we implemented InfoVis techniques such as Parallel Coordinates and risk signature charts to drive the decision process. As a case study, BrainXPlore offers a variety of information visualization modalities to present multivariate data in different ways. We evaluated BrainXPlore on a real dataset and accomplished acceptable results. The participating neurosurgeon gave us the feedback that BrainXPlore can decrease the time needed for biopsy planning and aid novice users in their decision making process.Additional Files and Images
Weblinks
No further information available.BibTeX
@mastersthesis{Pezenka-2016-MT,
title = "BrainXPlore - Decision finding in Brain Biopsy Planning",
author = "Lukas Pezenka",
year = "2017",
abstract = "Neurosurgeons make decisions based on expert knowledge that
takes factors such as safety margins, the avoidance of risk
structures, trajectory length and trajectory angle into
consideration. While some of those factors are mandatory,
others can be optimized in order to obtain the best possible
trajectory under the given circumstances. Through comparison
with the actually chosen trajectories from real biopsies and
qualitative interviews with domain experts, we identified
important rules for trajectory planning. In this thesis, we
present BrainXplore, an interactive visual analysis tool for
aiding neurosurgeons in planning brain biopsies. BrainXplore
is an extendable Biopsy Planning framework that incorporates
those rules while at the same time leaving full flexibility
for their customization and adding of new structures at
risk. Automatically computed candidate trajectories can be
incrementally refined in an interactive manner until an
optimal trajectory is found. We employ a spatial index
server as part of our system that allows us to access
distance information on an unlimited number of risk
structures at arbitrary resolution. Furthermore, we
implemented InfoVis techniques such as Parallel Coordinates
and risk signature charts to drive the decision process. As
a case study, BrainXPlore offers a variety of information
visualization modalities to present multivariate data in
different ways. We evaluated BrainXPlore on a real dataset
and accomplished acceptable results. The participating
neurosurgeon gave us the feedback that BrainXPlore can
decrease the time needed for biopsy planning and aid novice
users in their decision making process.",
month = dec,
address = "Favoritenstrasse 9-11/E193-02, A-1040 Vienna, Austria",
school = "Institute of Computer Graphics and Algorithms, Vienna
University of Technology ",
URL = "https://www.cg.tuwien.ac.at/research/publications/2017/Pezenka-2016-MT/",
}
 image
image Master Thesis
Master Thesis