Information
- Publication Type: Master Thesis
- Workgroup(s)/Project(s):
- Date: April 2017
- Date (Start): 1. June 2016
- Date (End): 21. April 2017
- First Supervisor: Ivan Viola
Abstract
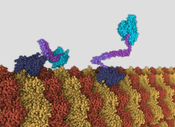
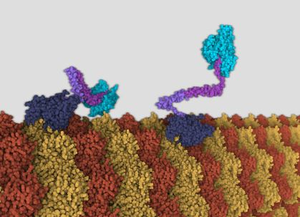
This work proposes an agent-based model for animating molecular machines. Usually molecular machines are visualized using key-frame animation. Creating large molecular assemblies with key-frame animation in standard 3D software can be a tedious task, because hundreds or thousands of molecular particles have to be animated by hand, considering various biological phenomena. To avoid repetitive animation of molecular particles, a prototypic framework is implemented, that employs an agent-based approach. Instead of animating the molecular particles directly, the framework utilizes behavior descriptions for each type of molecular particle. The animation results from the molecular particles interacting with each other as defined by their behavior. Interaction between molecular particles is enabled by an abstract model that is implemented by the framework. The methodology for creating the framework was driven through learning by example. Three molecular machines are visualized using the framework. During this process, the framework was iteratively improved, to meet the requirements for each new molecular machine. The resulted animations demonstrate that agent-based animation is a viable option for molecular machines.Additional Files and Images
Weblinks
No further information available.BibTeX
@mastersthesis{Gehrer-2017,
title = "Visualization of molecular machinery using agent-based
animation",
author = "Daniel Gehrer",
year = "2017",
abstract = "This work proposes an agent-based model for animating
molecular machines. Usually molecular machines are
visualized using key-frame animation. Creating large
molecular assemblies with key-frame animation in standard 3D
software can be a tedious task, because hundreds or
thousands of molecular particles have to be animated by
hand, considering various biological phenomena. To avoid
repetitive animation of molecular particles, a prototypic
framework is implemented, that employs an agent-based
approach. Instead of animating the molecular particles
directly, the framework utilizes behavior descriptions for
each type of molecular particle. The animation results from
the molecular particles interacting with each other as
defined by their behavior. Interaction between molecular
particles is enabled by an abstract model that is
implemented by the framework. The methodology for creating
the framework was driven through learning by example. Three
molecular machines are visualized using the framework.
During this process, the framework was iteratively improved,
to meet the requirements for each new molecular machine. The
resulted animations demonstrate that agent-based animation
is a viable option for molecular machines. ",
month = apr,
address = "Favoritenstrasse 9-11/E193-02, A-1040 Vienna, Austria",
school = "Institute of Computer Graphics and Algorithms, Vienna
University of Technology ",
URL = "https://www.cg.tuwien.ac.at/research/publications/2017/Gehrer-2017/",
}
 image
image Master thesis
Master thesis