Information
- Publication Type: Conference Paper
- Workgroup(s)/Project(s):
- Date: 2016
- Lecturer: Nicholas Waldin
- Event: VCBM
- Booktitle: Eurographics Workshop on Visual Computing for Biology and Medicine
Abstract
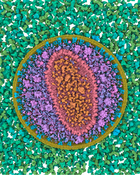
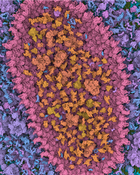
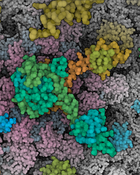
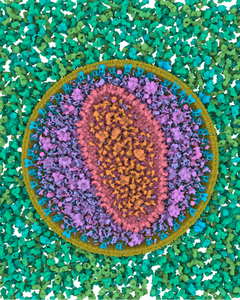
Visualization of structural biology data uses color to categorize or separate dense structures into particular semantic units. In multiscale models of viruses or bacteria, there are atoms on the finest level of detail, then amino-acids, secondary structures, macromolecules, up to the compartment level and, in all these levels, elements can be visually distinguished by color. However, currently only single scale coloring schemes are utilized that show information for one particular scale only. We present a novel technology which adaptively, based on the current scale level, adjusts the color scheme to depict or distinguish the currently best visible structural information. We treat the color as a visual resource that is distributed given a particular demand. The changes of the color scheme are seamlessly interpolated between the color scheme from the previous views into a given new one. With such dynamic multi-scale color mapping we ensure that the viewer is able to distinguish structural detail that is shown on any given scale. This technique has been tested by users with an expertise in structural biology and has been overall well received.Additional Files and Images
Weblinks
BibTeX
@inproceedings{Waldin_Nicholas_2016_Chameleon,
title = "Chameleon Dynamic Color Mapping for Multi-Scale Structural
Biology Models",
author = "Nicholas Waldin and Mathieu Le Muzic and Manuela Waldner and
Eduard Gr\"{o}ller and David Goodsell and Ludovic Autin and
Ivan Viola",
year = "2016",
abstract = "Visualization of structural biology data uses color to
categorize or separate dense structures into particular
semantic units. In multiscale models of viruses or bacteria,
there are atoms on the finest level of detail, then
amino-acids, secondary structures, macromolecules, up to the
compartment level and, in all these levels, elements can be
visually distinguished by color. However, currently only
single scale coloring schemes are utilized that show
information for one particular scale only. We present a
novel technology which adaptively, based on the current
scale level, adjusts the color scheme to depict or
distinguish the currently best visible structural
information. We treat the color as a visual resource that is
distributed given a particular demand. The changes of the
color scheme are seamlessly interpolated between the color
scheme from the previous views into a given new one. With
such dynamic multi-scale color mapping we ensure that the
viewer is able to distinguish structural detail that is
shown on any given scale. This technique has been tested by
users with an expertise in structural biology and has been
overall well received.",
event = "VCBM",
booktitle = "Eurographics Workshop on Visual Computing for Biology and
Medicine",
URL = "https://www.cg.tuwien.ac.at/research/publications/2016/Waldin_Nicholas_2016_Chameleon/",
}

 paper
paper