Information
- Publication Type: Conference Paper
- Workgroup(s)/Project(s):
- Date: May 2013
- ISBN: 978-80-223-3377-1
- Series: SCCG '13
- Publisher: ACM Publishing House
- Organization: Comenius University, Bratislava
- Location: Smolenice, Slovak Republic
- Lecturer: Julius Parulek
- Editor: Roman Durikovič, Holly Rushmeier
- Booktitle: SCCG 2013 - 29th Proceedings Spring conference on Computer Graphics
- Conference date: 1. May 2013 – 3. May 2013
- Pages: 120 – 127
- Keywords: Implicit Surfaces, Level-of-detail, Visualization of Molecular Surfaces
Abstract
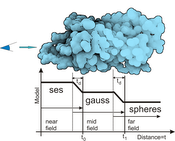
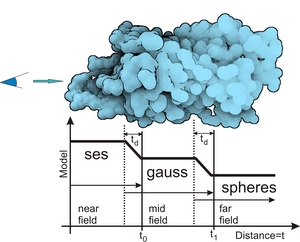
Molecular visualization is often challenged with rendering of large sequences of molecular simulations in real time. We introduce a novel approach that enables us to show even large protein complexes over time in real-time. Our method is based on the level-ofdetail concept, where we exploit three different molecular surface models, solvent excluded surface (SES), Gaussian kernels and van der Waals spheres combined in one visualization. We introduce three shading levels that correspond to their geometric counterparts and a method for creating seamless transition between these representations. The SES representation with full shading and added contours stands in focus while on the other side a sphere representation with constant shading and without contours provide the context. Moreover, we introduce a methodology to render the entire molecule directly using the A-buffer technique, which further improves the performance. The rendering performance is evaluated on series of molecules of varying atom counts.Additional Files and Images
Weblinks
No further information available.BibTeX
@inproceedings{Viola_Ivan_2013_SVA,
title = "Seamless Visual Abstraction of Molecular Surfaces",
author = "Julius Parulek and Timo Ropinski and Ivan Viola",
year = "2013",
abstract = "Molecular visualization is often challenged with rendering
of large sequences of molecular simulations in real time. We
introduce a novel approach that enables us to show even
large protein complexes over time in real-time. Our method
is based on the level-ofdetail concept, where we exploit
three different molecular surface models, solvent excluded
surface (SES), Gaussian kernels and van der Waals spheres
combined in one visualization. We introduce three shading
levels that correspond to their geometric counterparts and a
method for creating seamless transition between these
representations. The SES representation with full shading
and added contours stands in focus while on the other side a
sphere representation with constant shading and without
contours provide the context. Moreover, we introduce a
methodology to render the entire molecule directly using the
A-buffer technique, which further improves the performance.
The rendering performance is evaluated on series of
molecules of varying atom counts.",
month = may,
isbn = "978-80-223-3377-1",
series = " SCCG '13",
publisher = "ACM Publishing House",
organization = "Comenius University, Bratislava",
location = "Smolenice, Slovak Republic",
editor = "Roman Durikovi\v{c}, Holly Rushmeier",
booktitle = "SCCG 2013 - 29th Proceedings Spring conference on Computer
Graphics",
pages = "120--127",
keywords = "Implicit Surfaces, Level-of-detail, Visualization of
Molecular Surfaces",
URL = "https://www.cg.tuwien.ac.at/research/publications/2013/Viola_Ivan_2013_SVA/",
}

 Paper
Paper