Information
- Publication Type: Master Thesis
- Workgroup(s)/Project(s):
- Date: July 2009
- Diploma Examination: 20. July 2009
- First Supervisor:
Abstract
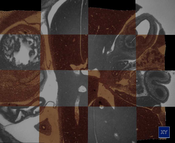
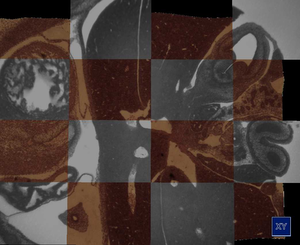
Weninger et al. [25] developed a novel methodology for rapid 2D and 3D computer analysis and visualization of gene expression patterns. The data is generated by staining a specimen followed by an iterating process of cutting thin slices and capturing them with an episcopic microscope. The result is an high resolution 3D dataset. One channel contains anatomical information and a second channel contains the gene expression patterns. In this thesis we examine methods for enhancing, registrating and visualizing this novel kind of data. We address the uneven illumination of slices that are introduced by the methodology. We developed an algorithm to fit a quadric surface through the background pixels to estimate the illumination situation over the whole slice. This estimate is used to correct the slices of one dataset. Further, an extension of this methodology was researched. Recycling the already cut sections for staining them a second time allows the medical domain scientists to augment their technique with additional information. The result of the second data generation phase is a stack of unaligned slices. The manual processing of the sections introduces non-linear deformations. We explored several registration algorithms to align the two image stacks. We found a two step registration approach to yield the best results. In the first step a coarse affine registration is used to approximately align the datasets. The result of the first step is inspected and if necessary corrected by the user. In the second step a b-spline registration is used that compensates for the non-linear deformations of the 2D slices. For the visual inspection of the registration results and to present an overview of the datasets we implemented two visualization approaches. A checkerboard view is used to compare 2D slices, and a three dimensional approach based on direct volume rendering incorporates surface enhancement by gradient magnitude opacity modulation to emphasize the alignment of tissue boundaries.Additional Files and Images
Weblinks
No further information available.BibTeX
@mastersthesis{brandorff-2009-erv,
title = "Enhancement, Registration, and Visualization of High
Resolution Episcopic Microscopy Data",
author = "Clemens Brandorff",
year = "2009",
abstract = "Weninger et al. [25] developed a novel methodology for rapid
2D and 3D computer analysis and visualization of gene
expression patterns. The data is generated by staining a
specimen followed by an iterating process of cutting thin
slices and capturing them with an episcopic microscope. The
result is an high resolution 3D dataset. One channel
contains anatomical information and a second channel
contains the gene expression patterns. In this thesis we
examine methods for enhancing, registrating and visualizing
this novel kind of data. We address the uneven illumination
of slices that are introduced by the methodology. We
developed an algorithm to fit a quadric surface through the
background pixels to estimate the illumination situation
over the whole slice. This estimate is used to correct the
slices of one dataset. Further, an extension of this
methodology was researched. Recycling the already cut
sections for staining them a second time allows the medical
domain scientists to augment their technique with additional
information. The result of the second data generation phase
is a stack of unaligned slices. The manual processing of the
sections introduces non-linear deformations. We explored
several registration algorithms to align the two image
stacks. We found a two step registration approach to yield
the best results. In the first step a coarse affine
registration is used to approximately align the datasets.
The result of the first step is inspected and if necessary
corrected by the user. In the second step a b-spline
registration is used that compensates for the non-linear
deformations of the 2D slices. For the visual inspection of
the registration results and to present an overview of the
datasets we implemented two visualization approaches. A
checkerboard view is used to compare 2D slices, and a three
dimensional approach based on direct volume rendering
incorporates surface enhancement by gradient magnitude
opacity modulation to emphasize the alignment of tissue
boundaries.",
month = jul,
address = "Favoritenstrasse 9-11/E193-02, A-1040 Vienna, Austria",
school = "Institute of Computer Graphics and Algorithms, Vienna
University of Technology ",
URL = "https://www.cg.tuwien.ac.at/research/publications/2009/brandorff-2009-erv/",
}
 image
image paper
paper