Overview
cellVIEW is an award-winning tool that provides fast rendering of very large biological macromolecular scenes and is inspired by state-of-the-art computer graphics techniques. cellVIEW is implemented in Unity3D, and it is publicly available at GitHub Open Source Repository. Because we utilize a game engine as development platform, we are able to release and deploy our tool faster, since our project only consists of a few scripts and shaders. We are then released from the burden of maintaining a complex software solution and from tedious deployment constraints. Additionally, since more people in the community are getting familiar with these engines, we ensure a maximum level of reproducibility among other researchers too, thus breaking the barriers caused by heterogeneous toolset usage across research departments. cellVIEW provides fast rendering by introducing new means to efficiently reduce the amount of processed geometries. CellVIEW is unique and has been specifically designed to match the ambitions of structural biologist to model and interactively visualize structures comprised of several billions atoms, which corresponds to sizes of small bacterial organisms.
The datasets that are displayed in cellVIEW were provided by cellPACK, a tool that populates and packs large biological models.
Application
cellVIEW is heavily dependent on the capabilities of your GPU. Your may have a more pleasant experience with cellVIEW if you stream it to your device and let the server do the computations. The current limit for one streaming session is 10 minutes.
Download
If you want to run cellVIEW locally you can download the lastest version here.
Awards
Publications
These academic publications describe several techniques and ideas that have influenced the development of cellVIEW.
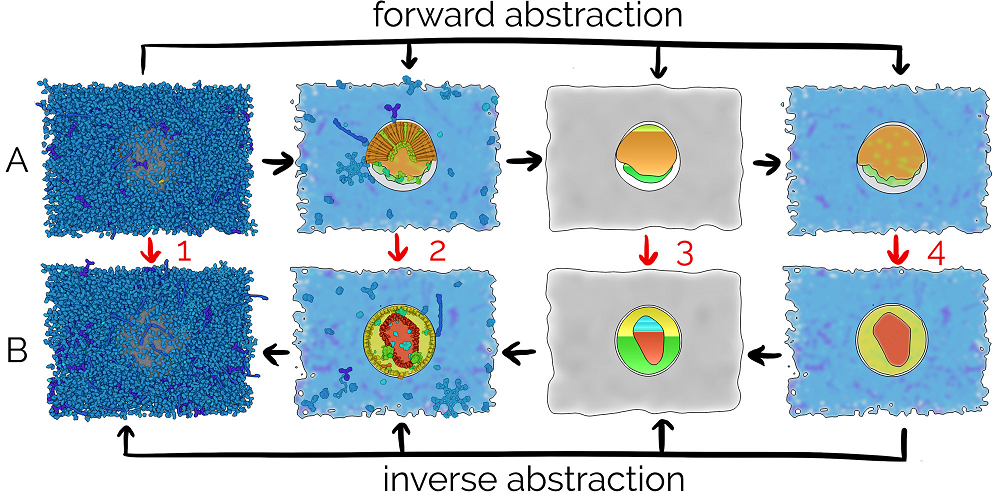
Illustrative Transitions in Molecular Visualization via Forward and Inverse Abstraction Transform
Johannes Sorger,
Peter Mindek,
Tobias Klein,
Graham Johnson,
Ivan Viola
In the 6th Eurographics Workshop on Visual Computing for Biology and Medicine (VCBM), 2016.
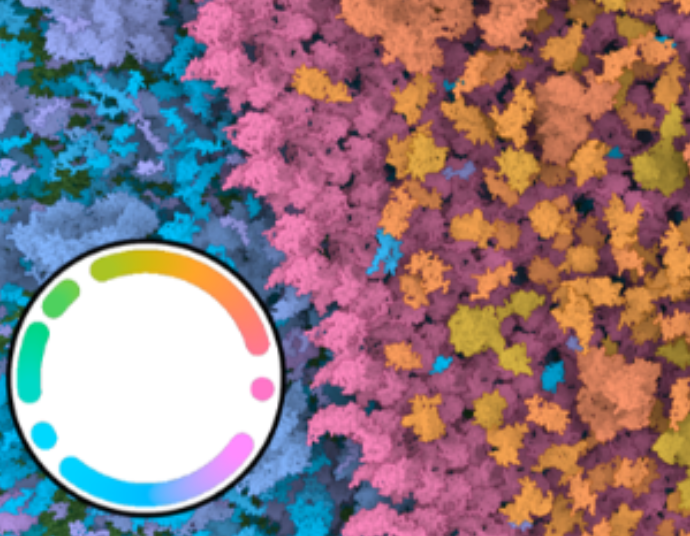
Chameleon - Dynamic Color Mapping for Multi-Scale Structural Biology Models
Nicholas Waldin,
Mathieu Le Muzic,
Manuela Waldner,
Eduard Gröller,
David Goodsell,
Ludovic Autin,
Ivan Viola
In the 6th Eurographics Workshop on Visual Computing for Biology and Medicine (VCBM), 2016.
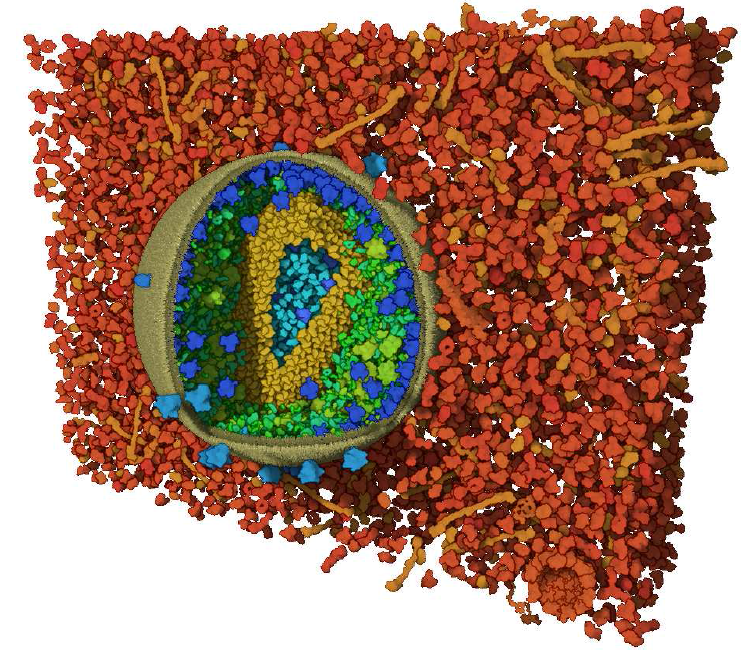
Visibility Equalizer: Cutaway Visualization of Mesoscopic Biological Models
Mathieu Le Muzic,
Peter Mindek,
Johannes Sorger,
Ludovic Autin,
David Goodsell,
Ivan Viola
Computer Graphics Forum, 35(3), 2016.

cellVIEW: a Tool for Illustrative and Multi-Scale Rendering of Large Biomolecular Datasets
Mathieu Le Muzic,
Ludovic Autin,
Julius Parulek,
Ivan Viola
In Eurographics Workshop on Visual Computing for Biology and Medicine, pages 61-70. September 2015.

Illustrative Timelapse: A Technique for Illustrative Visualization of Particle Simulations on the Mesoscale Level
Mathieu Le Muzic,
Manuela Waldner,
Julius Parulek,
Ivan Viola
In Visualization Symposium (PacificVis), 2015 IEEE Pacific, pages 247-254. April 2015.
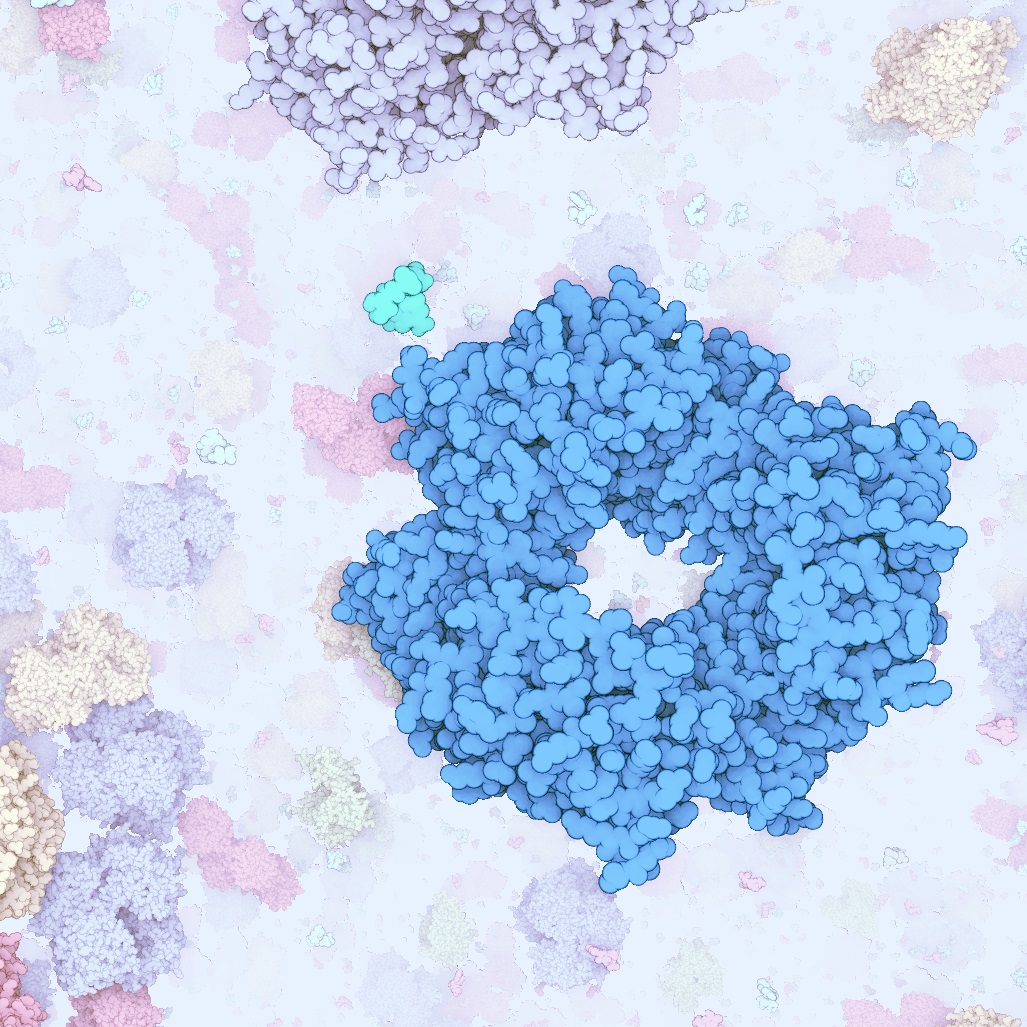
Illustrative Visualization of Molecular Reactions using Omniscient Intelligence and Passive Agents
Mathieu Le Muzic,
Julius Parulek,
Anne-Kristin Stavrum,
Ivan Viola
Computer Graphics Forum, 33(3):141-150, June 2014.
Research Team and Collaborators
Current Team (TU Wien)
Ivan Viola
Peter Mindek
Tobias Klein
Johannes Sorger
Nicholas Waldin
David Kouril
Mathieu Le Muzic
The Scripps Research Institute
Ludovic Autin
Arthur Olson
David Goodsell
Allen Institute for Cell Science
University of Bergen
Contact
For general questions of any kind please contact Ivan Viola.
