Abstract
Recent advances in medical imaging technology using multiple detector-row computed tomography (CT) provide volumetric datasets with unprecedented spatial resolution. This has allowed for CT to evolve into an excellent non-invasive vascular imaging technology, commonly referred to as CT-angiography. Visualization of vascular structures from CT datasets is demanding, however, and identification of anatomic objects in CT-datasets is highly desirable. Density and/or gradient operators have been used most commonly to classify CT data. In CT angiography, simple density/gradient operators do not allow precise and reliable classification of tissues due to the fact that different tissues (e.g. bones and vessels) possess the same density range and may lie in close spatial vicinity. We hypothesize, that anatomic classification can be achieved more accurately, if both spatial location and density properties of volume data are taken into account. We present a combination of two well-known methods for volume data processing to obtain accurate tissue classification. 3D watershed transform is used to partition the volume data in morphologically consistent blocks and a probabilistic anatomic atlas is used to distinguish between different kinds of tissues based on their density.Keywords:CT Angiography, Knowledge Based Segmentation Probabilistic Atlas, Thin-Plate-Spline, Histogram Classification
Download full paper
Matúš Straka, Alexandra La Cruz, Arnold Köchl, Miloš Šrámek, Eduard Gröller, Dominik Fleischmann "3D Watershed Transform Combined with a Probabilistic Atlas for Medical Image Segmentation", in proceedings of MIT2003, Full paper (642 kB).Figures in the paper
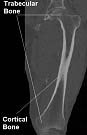 |
Figure 1: Different types of bone tissue (femur area) |
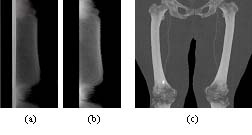 |
Figure 2: Dependency of density histograms on the slice position. Each row in (a) and (b) shows the histogram (1000HU to 3095HU) of the corresponding slice in a dataset, the MIP of which is presented in (c). (a) shows histograms of all body tissues while (b) only those of segmented bones. |
| Figure 3: (a) Vertical slice of original the dataset, (b) MIP of the bone probability volume (vessels are also enhanced and still present), (c) bone mask of (a) (vertical slice) obtained by region growing with help of probability atlas (vessels suppressed), (d) a MIP of (c). |
|
|
Figure 4: A vertical slices of a 3D dataset (hip joint area, cropped): (a) oversegmentation is a characteristic of the plain watershed transform (b) results of oversegmentation suppression achieved by merging regions according to their density mean values and (c) results of oversegmentation suppression achieved by both merging regions according to their density mean values and hierarchical merging |
|
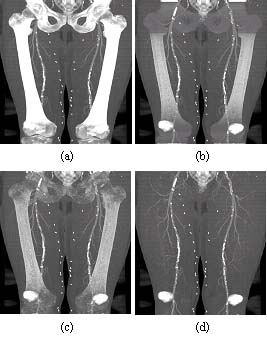 |
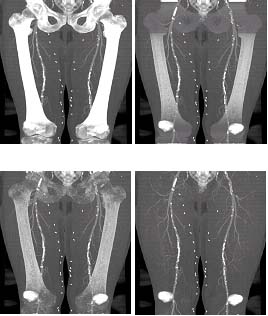
Figure 5: (a) MIP of the original dataset, (b) MIP, bone tissue partially removed using mask obtained with probabilistic atlas. (c) MIP, bone tissue removed using mask extended with 3D watershed transform, (d) MIP, bone tissue removed using the dilated mask. (Patella and sacro-coccygeal bone not modelled). |
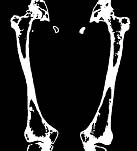 |
Figure 6: Bone mask extended by the 3D watershed region classification (compare to Figure 3c) |
|
Figure 7: (a) Final bone mask after dilation (vertical slice from dataset), (b) final bone model (surface shaded), without dilation, (c) for comparison: object mask acquired only with simple thresholding (th = 200HU), surface shaded. (Patella and sacro-coccygeal bones were not modelled) |
BibTeX Entry
@InProceedings{Straka:03:3DW,
author = "Matúš Straka and Alexandra La Cruz and
Arnold Köchl and Miloš Šrámek and Dominik Fleischmann and
Eduard Gröller",
title = "3D Watershed Transform Combined with a
Probabilistic Atlas for Medical Image Segmentation",
bookTitle = "Proceedings of MIT 2003",
year = "2003",
pages = "1--8",
keywords = "CT Angiography, Knowledge Based Segmentation,
Probabilistic Atlas, Thin-Plate-Spline, Histogram Classification"
}