Information
- Publication Type: Journal Paper with Conference Talk
- Workgroup(s)/Project(s):
- Date: January 2016
- Journal: IEEE Transactions on Visualization and Computer Graphics
- Volume: 22
- Number: 1
- Location: Illinois, Chicago, USA
- Lecturer: Jan Byska
- ISSN: 1077-2626
- Event: IEEE Vis 2015
- Conference date: 25. October 2015 – 30. October 2015
- Pages: 747 – 756
- Keywords: aggregation, molecular dynamics, Protein, interaction, tunnel
Abstract
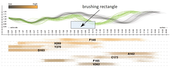
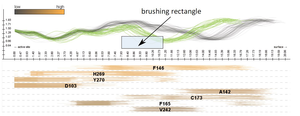
In this paper we propose a novel method for the interactive exploration of protein tunnels. The basic principle of our approach is that we entirely abstract from the 3D/4D space the simulated phenomenon is embedded in. A complex 3D structure and its curvature information is represented only by a straightened tunnel centerline and its width profile. This representation focuses on a key aspect of the studied geometry and frees up graphical estate to key chemical and physical properties represented by surrounding amino acids. The method shows the detailed tunnel profile and its temporal aggregation. The profile is interactively linked with a visual overview of all amino acids which are lining the tunnel over time. In this overview, each amino acid is represented by a set of colored lines depicting the spatial and temporal impact of the amino acid on the corresponding tunnel. This representation clearly shows the importance of amino acids with respect to selected criteria. It helps the biochemists to select the candidate amino acids for mutation which changes the protein function in a desired way. The AnimoAminoMiner was designed in close cooperation with domain experts. Its usefulness is documented by their feedback and a case study, which are included.Additional Files and Images
Weblinks
BibTeX
@article{Viola_Ivan_2015_AAM,
title = "AnimoAminoMiner: Exploration of Protein Tunnels and their
Properties in Molecular Dynamics",
author = "Jan Byska and Mathieu Le Muzic and Eduard Gr\"{o}ller and
Ivan Viola and Barbora Kozlikova",
year = "2016",
abstract = "In this paper we propose a novel method for the interactive
exploration of protein tunnels. The basic principle of our
approach is that we entirely abstract from the 3D/4D space
the simulated phenomenon is embedded in. A complex 3D
structure and its curvature information is represented only
by a straightened tunnel centerline and its width profile.
This representation focuses on a key aspect of the studied
geometry and frees up graphical estate to key chemical and
physical properties represented by surrounding amino acids.
The method shows the detailed tunnel profile and its
temporal aggregation. The profile is interactively linked
with a visual overview of all amino acids which are lining
the tunnel over time. In this overview, each amino acid is
represented by a set of colored lines depicting the spatial
and temporal impact of the amino acid on the corresponding
tunnel. This representation clearly shows the importance of
amino acids with respect to selected criteria. It helps the
biochemists to select the candidate amino acids for mutation
which changes the protein function in a desired way. The
AnimoAminoMiner was designed in close cooperation with
domain experts. Its usefulness is documented by their
feedback and a case study, which are included.",
month = jan,
journal = "IEEE Transactions on Visualization and Computer Graphics",
volume = "22",
number = "1",
issn = "1077-2626",
pages = "747--756",
keywords = "aggregation, molecular dynamics, Protein, interaction,
tunnel",
URL = "https://www.cg.tuwien.ac.at/research/publications/2016/Viola_Ivan_2015_AAM/",
}

 Paper
Paper