Information
- Publication Type: Master Thesis
- Workgroup(s)/Project(s):
- Date: August 2013
- TU Wien Library:
- First Supervisor:
- Eduard Gröller
- Hendrik Schulze
- Eduard Gröller
Abstract
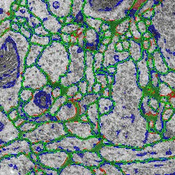
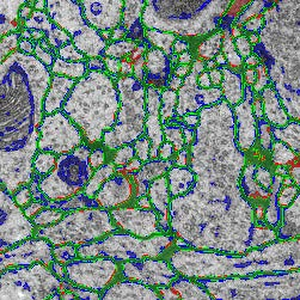
In order to (preferably) automatically derive the neuronal structures from brain tissue image stacks, the research field computational neuroanatomy relies on computer assisted techniques such as visualization, machine learning and analysis. The image acquisition is based on the so-called transmission electron microscopy (TEM) that allows resolution that is high enough to identify relevant structures in brain tissue images (less than 5 nm per pixel). In order to get to an image stack (or volume) the tissue samples are sliced (or sectioned) with a diamond knife in slices of 40 nm thickness. This approach is called serial-section transmission electron microscopy (ssTEM). The manual segmentation of these high-resolution, low-contrast and artifact afflicted images would be impracticable alone due to the high resolution of 200,000 images of size 2m x 2m pixel in a cubic centimeter tissue sample. But, the automatic segmentation is error-prone due to the small pixel value range (8 bit per pixel) and diverse artifacts resulting from mechanical sectioning of tissue samples. Additionally, the biological samples in general contain densely packed structures which leads to non-uniform background that introduces artifacts as well. Therefore, it is important to quantify, visualize and reproduce the automatic segmentation results interactively with as few user interaction as possible.This thesis is based on the membrane segmentation proposed by Kaynig-Fittkau [2011] which for ssTEM brain tissue images outputs two results: (a) a certainty value per pixel (with regard to the analytical model of the user selection of cell membrane pixels) which states how certain the underlying statistical model is that the pixel is belonging to the membrane , and (b) after an optimization step the resulting edges which represent the membrane. In this work we present a visualization-assisted method to explore the parameters of the segmentation. The aim is to interactively mark those regions where the segmentation fails to the expert user in order to structure the post- or re-segmentation or to prove-read the segmentation results. This is achieved by weighting the membrane pixels by the uncertainty values resulting from the segmentation process.
We would like to start here and employ user knowledge once more to decide which data and in what form should be introduced to the random forest classifier in order to improve the segmentation results either through segmentation quality or segmentation speed. In this regard we use focus our attention especially on the visualizations of the uncertainty, the error and multi-modal data. The interaction techniques are explicitly used in those cases where we expect the highest gain at the end of the exploration. We show the effectiveness of the proposed methods using the freely available ssTEM brain tissue dataset of the drosophila fly. Because we lack the expert knowledge in the field of neuroanatomy re must rely our assumptions and methods on the underlying ground truth segmentations of the drosophila fly brain tissue dataset.
Additional Files and Images
Weblinks
No further information available.BibTeX
@mastersthesis{Maricic_2013_VFE,
title = "Visual Feature Exploration for ssTEM Image Segmentation",
author = "Ivan Maricic",
year = "2013",
abstract = " In order to (preferably) automatically derive the neuronal
structures from brain tissue image stacks, the research
field computational neuroanatomy relies on computer assisted
techniques such as visualization, machine learning and
analysis. The image acquisition is based on the so-called
transmission electron microscopy (TEM) that allows
resolution that is high enough to identify relevant
structures in brain tissue images (less than 5 nm per
pixel). In order to get to an image stack (or volume) the
tissue samples are sliced (or sectioned) with a diamond
knife in slices of 40 nm thickness. This approach is called
serial-section transmission electron microscopy (ssTEM). The
manual segmentation of these high-resolution, low-contrast
and artifact afflicted images would be impracticable alone
due to the high resolution of 200,000 images of size 2m x 2m
pixel in a cubic centimeter tissue sample. But, the
automatic segmentation is error-prone due to the small pixel
value range (8 bit per pixel) and diverse artifacts
resulting from mechanical sectioning of tissue samples.
Additionally, the biological samples in general contain
densely packed structures which leads to non-uniform
background that introduces artifacts as well. Therefore, it
is important to quantify, visualize and reproduce the
automatic segmentation results interactively with as few
user interaction as possible. This thesis is based on the
membrane segmentation proposed by Kaynig-Fittkau [2011]
which for ssTEM brain tissue images outputs two results: (a)
a certainty value per pixel (with regard to the analytical
model of the user selection of cell membrane pixels) which
states how certain the underlying statistical model is that
the pixel is belonging to the membrane , and (b) after an
optimization step the resulting edges which represent the
membrane. In this work we present a visualization-assisted
method to explore the parameters of the segmentation. The
aim is to interactively mark those regions where the
segmentation fails to the expert user in order to structure
the post- or re-segmentation or to prove-read the
segmentation results. This is achieved by weighting the
membrane pixels by the uncertainty values resulting from the
segmentation process. We would like to start here and
employ user knowledge once more to decide which data and in
what form should be introduced to the random forest
classifier in order to improve the segmentation results
either through segmentation quality or segmentation speed.
In this regard we use focus our attention especially on the
visualizations of the uncertainty, the error and multi-modal
data. The interaction techniques are explicitly used in
those cases where we expect the highest gain at the end of
the exploration. We show the effectiveness of the proposed
methods using the freely available ssTEM brain tissue
dataset of the drosophila fly. Because we lack the expert
knowledge in the field of neuroanatomy re must rely our
assumptions and methods on the underlying ground truth
segmentations of the drosophila fly brain tissue dataset. ",
month = aug,
address = "Favoritenstrasse 9-11/E193-02, A-1040 Vienna, Austria",
school = "Institute of Computer Graphics and Algorithms, Vienna
University of Technology ",
URL = "https://www.cg.tuwien.ac.at/research/publications/2013/Maricic_2013_VFE/",
}
 Poster
Poster Thesis
Thesis