Introduction
The structure of molecules can be complex and hard to understand. In order to tackle these problems molecular surface maps can be used to represent the whole structure of a 3D molecule onto a 2D surface. With this approach it is much easier for the analyst to understand the molecular surface. Arbitrary attributes of the original molecular surface, such as biochemical properties or geometrical features, can be identified from the 2D representation of the surface. Analysts can assess all molecular surface attributes from this texture. The implementation of the program is based on the research from [Krone et al. 2016].
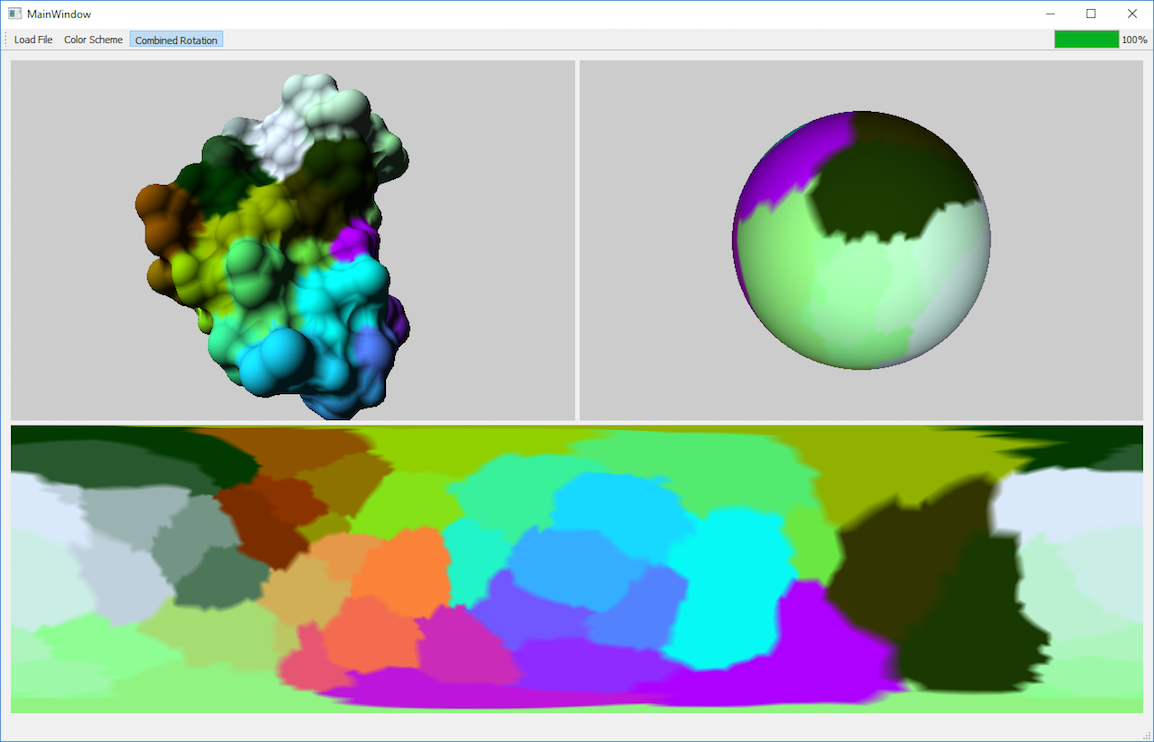
The Program
There is a 'Load File' button in the menu bar of the program which is used to load molecules from the [Protein Data Bank] into the program. Once this is done, the mesh of the molecule should be visible in the top left window. With a drag of the mouse, the mesh can be rotated. The top right window is used to display the sphere with the molecule surface map. If the molecule or the sphere is rotated, both view rotation together. This helps to get a better understanding of the mapping. The button 'Color Scheme' which is also located in the menu bar, can be used to change the colors used for the molecule. This is helpful to highlight different qualities.
Binarys Windows: [Download]
Source Code: [Download]
Technologies Used
- Programming Language: C++
- Framework: Qt
- Graphic API: OpenGL
 1.8.14
1.8.14