Overview
The Molecule Visualizer is a program to visualize molecules in a real-time 3D application.
The user can load a molecule file from the
Protein Data Bank with PDB format.
The challenge when visualizing molecules is to find ways to improve the perception of the spatial structure. Especially large molecules are problematic.
The methods that were chosen to tackle the perception problem are described in the "Functionality" section. A more detailed description can be found in the paper
"Ambient Occlusion and Edge Cueing to Enhance Real Time Molecular Visualization" by Tarini et al. [1]. The implementation is based on the algorithms presented in that paper.
The program was implemented as an assignment of the course "Visualization 2" at the Technical University of Vienna in the summer semester 2014.
To Top
Getting Started
Download the zipped files. Extract the content of the folder and run the executable.
Make sure that the necessary OpenGL and glew dlls are located where Windows can find them. The program should look like this:
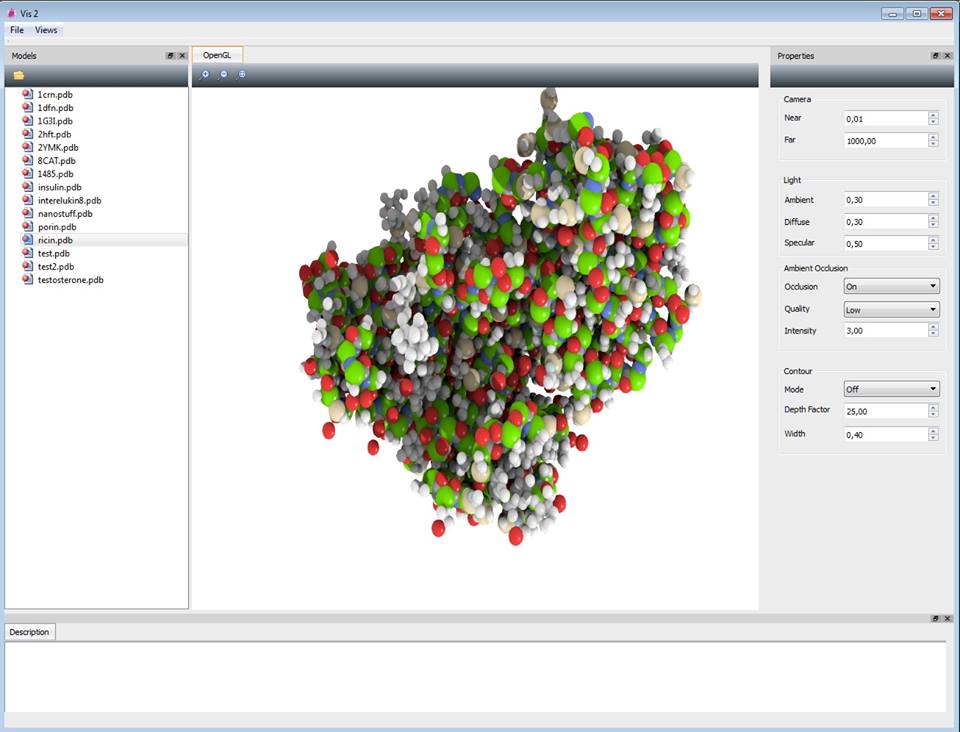
On the left side of the window there are some sample models that can be loaded immediately. The main view shows the visualization and buttons to zoom in, zoom out and reset to defaults.
On the right side there is a settings section, where parameters can be changed. The modifications are applied in real-time. Only the initial load of the molecule may take some time, since the ambient occlusion is precomputed.
To Top
Functionality
Three effects are implemented to improve the perception of the molecule: ambient occlusion, depth-aware contours and percentage-closer soft shadows.
Two of these are presented in the paper [1]: ambient occlusion and depth-aware contours.
Ambient occlusion
According to the chosen ambient occlusion quality a set of sampling directions is precomputed. It may either consist of 66, 162 or 642 directions.
For each direction an off-screen rendering pass is performed. In the first pass a depth map is created. The next pass writes over the texture
for the molecule and accesses the depth map produced in the previous pass.
For each primitive a quad covering the corresponding texture patch is sent, together with attributes encoding the position and shape of the primitive.
For every fragment at position
(u, v) a point
p in 3D space is computed, which then is transformed with the same viewing conditions used
in the first pass. The resulting depth is compared with the one extracted from the depth map to determine if the point
p is lit by light coming from
the current viewing direction. If so, the light contribution is accumulated at the corresponding pixel through alpha blending.
Depth-aware contours
When using the dynamic depth-aware contour lines, the width of the contours around the spheres depends on the depth difference to the next sphere behind the current one.
If there is no further sphere the maximal width is used.
If two spheres interset no contour will be rendered at the intersection.
Since impostor-based rendering is used it is fairly simple to add depth-aware contours.
The size of the 2D quads is increased and drawn as a border with a given width.
All pixels with a certain distance to the center of the current sphere belong to the contour and are drawn as black pixels.
To make the contour depth aware, the contour pixels are also moved to the back (depth increased) depending on the
distance to the border of the sphere: The further away from the sphere border, the further the contour pixel is moved to the back.
To Top
Test Environment
Intel Core i5 2.6 GHz
Nvidia NVS 5400m
8 GB RAM
To Top
References
[1] M. Tarini, P. Cignoni and C. Montani. Ambient Occlusion and Edge Cueing to Enhance Real Time Molecular Visualization. In
IEEE Transactions on Visualization and Computer Graphics. Vol. 12. No. 5. 2006.
[2] Background Image from "Free picture": (
molecule on
Free desktop pictures)
To Top
 On the left side of the window there are some sample models that can be loaded immediately. The main view shows the visualization and buttons to zoom in, zoom out and reset to defaults.
On the right side there is a settings section, where parameters can be changed. The modifications are applied in real-time. Only the initial load of the molecule may take some time, since the ambient occlusion is precomputed.
To Top
On the left side of the window there are some sample models that can be loaded immediately. The main view shows the visualization and buttons to zoom in, zoom out and reset to defaults.
On the right side there is a settings section, where parameters can be changed. The modifications are applied in real-time. Only the initial load of the molecule may take some time, since the ambient occlusion is precomputed.
To Top
