Information
- Publication Type: Journal Paper (without talk)
- Workgroup(s)/Project(s):
- Date: November 2013
- ISSN: 1471-2105
- Journal: BMC Bioinformatics
- Number: Suppl 19:S4
- Volume: 14
- Pages: 1 – 15
Abstract
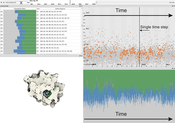
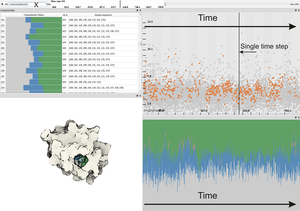
Molecular surfaces provide a useful mean for analyzing interactions between biomolecules; such as identification and characterization of ligand binding sites to a host macromolecule. We present a novel technique, which extracts potential binding sites, represented by cavities, and characterize them by 3D graphs and by amino acids. The binding sites are extracted using an implicit function sampling and graph algorithms. We propose an advanced cavity exploration technique based on the graph parameters and associated amino acids. Additionally, we interactively visualize the graphs in the context of the molecular surface. We apply our method to the analysis of MD simulations of Proteinase 3, where we verify the previously described cavities and suggest a new potential cavity to be studied.Additional Files and Images
Weblinks
BibTeX
@article{Viola_Ivan_2013_VCA,
title = "Visual cavity analysis in molecular simulations",
author = "Julius Parulek and Cagatay Turkay and Nathalie Reuter and
Ivan Viola",
year = "2013",
abstract = "Molecular surfaces provide a useful mean for analyzing
interactions between biomolecules; such as identification
and characterization of ligand binding sites to a host
macromolecule. We present a novel technique, which extracts
potential binding sites, represented by cavities, and
characterize them by 3D graphs and by amino acids. The
binding sites are extracted using an implicit function
sampling and graph algorithms. We propose an advanced cavity
exploration technique based on the graph parameters and
associated amino acids. Additionally, we interactively
visualize the graphs in the context of the molecular
surface. We apply our method to the analysis of MD
simulations of Proteinase 3, where we verify the previously
described cavities and suggest a new potential cavity to be
studied.",
month = nov,
issn = "1471-2105",
journal = "BMC Bioinformatics",
number = "Suppl 19:S4 ",
volume = "14",
pages = "1--15",
URL = "https://www.cg.tuwien.ac.at/research/publications/2013/Viola_Ivan_2013_VCA/",
}

 Paper
Paper