| Image |
Bib Reference |
Publication Type |
| 2019 |
 |
Maximilian Sbardellati, Haichao Miao, Hsiang-Yun Wu , Eduard Gröller , Eduard Gröller , Ivan Barisic, Ivan Viola , Ivan Barisic, Ivan Viola
Interactive Exploded Views for Molecular Structures
In Proceedings of the 9th Eurographics Workshop on Visual Computing for Biology and Medicine, pages 103-112. September 2019.
[ paper] paper] |
Conference Paper |
 |
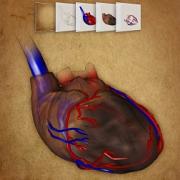
Peter Mindek, Tobias Klein, Ludovic Autin, Theresia Gschwandtner
Microtubule Catastrophe
VCBM 2019, Image Contest Jury's Award
[ Microtubule Catastrophe] Microtubule Catastrophe] |
Miscellaneous Publication |
 |
Robert Horvath
Image-Space Metaballs Using Deep Learning
[ image] [ image] [ Master Thesis] Master Thesis] |
Master Thesis |
 |
Pascal Plank
Effective Line Drawing Generation
[ image] [ image] [ Master Thesis] Master Thesis] |
Master Thesis |
 |
Hsiang-Yun Wu , Martin Nöllenburg, Filipa L. Sousa, Ivan Viola , Martin Nöllenburg, Filipa L. Sousa, Ivan Viola
Metabopolis: Scalable Network Layout for Biological Pathway Diagrams in Urban Map Style
BMC Bioinformatics, 20(187):1-20, May 2019. [ paper] [video] paper] [video] |
Journal Paper (without talk) |
 |
Eric Mörth
Interactive Reformation of Fetal Ultrasound Data to a T-Position
[ image] [ image] [ Master Thesis] Master Thesis] |
Master Thesis |
| 2018 |
 |
Peter Mindek
Molecular Visualization,
10. November 2018, FYI: Informationsdesign Konferenz Wien, Nordbahn-Halle, Vienna
|
WorkshopTalk |
 |
Thomas Bernhard Koch
Semantic Screen-Space Occlusion for Multiscale Molecular Visualization
[ Bachelor Thesis] Bachelor Thesis] |
Bachelor Thesis |
 |
Lukas Lipp
Visualization of Fiber Orientation in Glass Fiber Reinforced Polymers
[ Bachelor Thesis] Bachelor Thesis] |
Bachelor Thesis |
 |
Haichao Miao, Elisa De Llano, Johannes Sorger, Yasaman Ahmadi, Tadija Kekic, Tobias Isenberg, Eduard Gröller , Ivan Barisic, Ivan Viola , Ivan Barisic, Ivan Viola
Multiscale Visualization and Scale-adaptive Modification of DNA Nanostructures
IEEE Transactions on Visualization and Computer Graphics, 24(1), January 2018. [ paper] paper] |
Journal Paper with Conference Talk |
 |
Stefan Dietrich
Smart Visibility Technique for Linear and Planar Molecular Assemblies
|
Bachelor Thesis |
| 2017 |
|
Nicholas Waldin
Using and Adapting to Limits of Human Perception in Visualization
Supervisor: Ivan Viola
[ thesis] thesis] |
PhD-Thesis |
 |
Peter Mindek, Gabriel Mistelbauer, Eduard Gröller , Stefan Bruckner , Stefan Bruckner
Data-Sensitive Visual Navigation
Computers & Graphics, 67(C):77-85, October 2017. [ Paper] Paper] |
Journal Paper with Conference Talk |
 |
Ivan Viola , Martin Seyfert , Martin Seyfert
Dynamic word clouds
In Proceedings of SCCG 2017. May 2017.
[ image] [ image] [ paper] paper] |
Conference Paper |
 |
Daniel Gehrer, Ivan Viola
Visualization of molecular machinery using agent-based animation
In Proceedings of SCCG 2017. May 2017.
[ paper] paper] |
Conference Paper |
|
Nicholas Waldin, Manuela Waldner , Ivan Viola , Ivan Viola
Flicker Observer Effect: Guiding Attention Through High Frequency Flicker in Images
Computer Graphics Forum, 36(2):467-476, May 2017. [ paper] paper] |
Journal Paper with Conference Talk |
 |
David Kouřil
Maya2CellVIEW: 3D Package Integrated Tool for Creating Large and Complex Molecular Scenes
|
Master Thesis |
 |
Haichao Miao, Gabriel Mistelbauer, Alexey Karimov, Amir Alansary, Alice Davidson, David F.A. Lloyd, Mellisa Damodaram, Lisa Story, Jana Hutter, Joseph V. Hajnal, Mary Rutherford, Bernhard Preim, Bernhard Kainz, Eduard Gröller
Placenta Maps: In Utero Placental Health Assessment of the Human Fetus
IEEE Transactions on Visualization and Computer Graphics, 23(6):1612-1623, 2017. |
Journal Paper with Conference Talk |
 |
Peter Mindek, David Kouřil, Johannes Sorger, David Toloudis, Blair Lyons, Graham Johnson, Eduard Gröller , Ivan Viola , Ivan Viola
Visualization Multi-Pipeline for Communicating Biology
IEEE Transactions on Visualization and Computer Graphics, 24(1), 2017. [ Paper] [ Paper] [ Preview Movie] [ Preview Movie] [ Slides] Slides] |
Journal Paper with Conference Talk |
| 2016 |
 |
Johannes Sorger, Peter Mindek, Tobias Klein, Graham Johnson, Ivan Viola
Illustrative Transitions in Molecular Visualization via Forward and Inverse Abstraction Transform
In Eurographics Workshop on Visual Computing for Biology and Medicine (VCBM), pages 21-30. September 2016.
[ paper] [ paper] [ video] video] |
Conference Paper |
 |
Lucas Dworschak
Semantically Zoomable Choropleth Map
|
Bachelor Thesis |
 |
Matthias Bernhard, Manuela Waldner , Pascal Plank, Veronika Solteszova, Ivan Viola , Pascal Plank, Veronika Solteszova, Ivan Viola
The Accuracy of Gauge-Figure Tasks in Monoscopic and Stereo Displays
IEEE Computer Graphics and Applications, 36(4):56-66, July 2016. |
Journal Paper (without talk) |
 |
Hani Gadllah
Comparative Visualization of the Circle of Willis
[ image] image] |
Bachelor Thesis |
 |
Peter Mindek
Multi-Scale Molecular Data Visualization,
3. May 2016, QCB Workshop on Visualizing & Modeling Cell Biology, Salt Lake City, Utah, USA
[ Slides] Slides] |
WorkshopTalk |
|
Maximilian Langer
Dynamic Multiscale Vector Volumes
[ Thesis] Thesis] |
Bachelor Thesis |
 |
Jan Byska, Mathieu Le Muzic, Eduard Gröller , Ivan Viola , Ivan Viola , Barbora Kozlikova , Barbora Kozlikova
AnimoAminoMiner: Exploration of Protein Tunnels and their Properties in Molecular Dynamics
IEEE Transactions on Visualization and Computer Graphics, 22(1):747-756, January 2016. [ Paper] Paper] |
Journal Paper with Conference Talk |
 |
Michael Krone, Barbora Kozlikova, Norbert Lindow, Marc Baaden, Daniel Baum, Julius Parulek, Hans-Christian Hege, Ivan Viola
Visual Analysis of Biomolecular Cavities: State of the Art
Computer Graphics Forum, 35(3):527-551, 2016. [ paper] paper] |
Journal Paper with Conference Talk |
 |
Ivan Viola
Effective Visual Representations
[ thesis] thesis] |
Habilitation Thesis |
|
Matthias Reisacher
CellPathway a Simulation Tool for Illustrative Visualization of Biochemical Networks
[ Thesis] Thesis] |
Bachelor Thesis |
 |
Matthias Reisacher, Mathieu Le Muzic, Ivan Viola
CellPathway: A Simulation Tool for Illustrative Visualization of Biochemical Networks
In Proceedings of WSCG. 2016.
[ paper] paper] |
Conference Paper |
 |
Veronika Solteszova, Åsmund Birkeland, Sergej Stoppel, Ivan Viola , Stefan Bruckner , Stefan Bruckner
Output-Sensitive Filtering of Streaming Volume Data
Computer Graphics Forum, 35, 2016. [ paper] paper] |
Journal Paper (without talk) |
 |
Daniel Cornel, Artem Konev, Berhard Sadransky, Zsolt Horvath, Andrea Brambilla, Ivan Viola , Jürgen Waser , Jürgen Waser
Composite Flow Maps
Computer Graphics Forum, 35(3):461-470, 2016. [ paper] paper] |
Journal Paper with Conference Talk |
 |
Matthias Glinzner
Texturing of 3D Objects using Simple Physics and Equilateral Triangle Patches
[ paper] paper] |
Bachelor Thesis |
|
Viktor Vad, Douglas Cedrim, Wolfgang Busch, Peter Filzmoser, Ivan Viola
Generalized box-plot for root growth ensembles
BMC Bioinformatics, 2016. |
Journal Paper with Conference Talk |
 |
Mathieu Le Muzic, Peter Mindek, Johannes Sorger, Ludovic Autin, David Goodsell, Ivan Viola
Visibility Equalizer: Cutaway Visualization of Mesoscopic Biological Models
Computer Graphics Forum, 35(3), 2016. [ Paper] [ Paper] [ Slides] Slides] |
Journal Paper with Conference Talk |
| 2015 |
|
Alexandra Diehl , Leandro Pelorosso, Kresimir Matkovic, Claudio Delrieux, Marc Ruiz, Eduard Gröller , Leandro Pelorosso, Kresimir Matkovic, Claudio Delrieux, Marc Ruiz, Eduard Gröller , Stefan Bruckner , Stefan Bruckner
Albero: A Visual Analytics Tool for Probabilistic Weather Forecasting.
Poster shown at Poster at Workshop Big Data & Environment
(November 2015)
|
Poster |
 |
Mathieu Le Muzic, Ludovic Autin, Julius Parulek, Ivan Viola
cellVIEW: a Tool for Illustrative and Multi-Scale Rendering of Large Biomolecular Datasets
In Eurographics Workshop on Visual Computing for Biology and Medicine, pages 61-70. September 2015.
[ ] [ ] [ paper] paper] |
Conference Paper |
 |
Pascal Plank
Human Visual Perception of 3D Surfaces
|
Bachelor Thesis |
 |
Barbora Kozlikova, Michael Krone, Norbert Lindow, Martin Falk, Marc Baaden, Daniel Baum, Ivan Viola , Julius Parulek, Hans-Christian Hege , Julius Parulek, Hans-Christian Hege
Visualization of Biomolecular Structures: State of the Art
In Eurographics Conference on Visualization (EuroVis) - STARs, pages 061-081. May 2015.
[ Paper] Paper] |
Conference Paper |
 |
Jan Byska, Adam Jurcik, Eduard Gröller , Ivan Viola , Ivan Viola , Barbora Kozlikova , Barbora Kozlikova
MoleCollar and Tunnel Heat Map Visualizations for Conveying Spatio-Temporo-Chemical Properties Across and Along Protein Voids
Computer Graphics Forum, 3(34):1-10, May 2015. [ Paper] Paper] |
Journal Paper with Conference Talk |
 |
Mathieu Le Muzic, Manuela Waldner , Julius Parulek, Ivan Viola , Julius Parulek, Ivan Viola
Illustrative Timelapse: A Technique for Illustrative Visualization of Particle Simulations on the Mesoscale Level
In Visualization Symposium (PacificVis), 2015 IEEE Pacific, pages 247-254. April 2015.
[ paper] [ paper] [ screenshot] screenshot] |
Conference Paper |
 |
Peter Mindek, Ladislav Čmolík, Ivan Viola , Eduard Gröller , Eduard Gröller , Stefan Bruckner , Stefan Bruckner
Automatized Summarization of Multiplayer Games
In Proceedings of Spring Conference on Computer Graphics 2015, pages 93-100. April 2015.
[ Paper] [ Paper] [ Slides] Slides] |
Conference Paper |
 |
Simon Brenner
Projector-Based Textures for 3D-Printed Models - Tangible Molecular Visualization
|
Master Thesis |
| 2014 |
 |
Manuela Waldner , Mathieu Le Muzic, Matthias Bernhard, Werner Purgathofer , Mathieu Le Muzic, Matthias Bernhard, Werner Purgathofer , Ivan Viola , Ivan Viola
Attractive Flicker: Guiding Attention in Dynamic Narrative Visualizations
IEEE Transactions on Visualization and Computer Graphics, 20(12):2456-2465, December 2014. [ paper] [ paper] [ Preview video] [ Preview video] [ Submission video] Submission video] |
Journal Paper with Conference Talk |
 |
Roman Püngüntzky2
HistoryTime: A Chrome history visualization using WebGL
[ thesis] thesis] |
Bachelor Thesis |
 |
Åsmund Birkeland, Cagatay Turkay, Ivan Viola
Perceptually Uniform Motion Space
IEEE Transactions on Visualization and Computer Graphics, 20(11):1542-1554, November 2014. |
Journal Paper with Conference Talk |
 |
Ivan Kolesár, Julius Parulek, Ivan Viola , Stefan Bruckner , Stefan Bruckner , Anne-Kristin Stavrum, Helwig Hauser , Anne-Kristin Stavrum, Helwig Hauser
Interactively illustrating polymerization using three-level model fusion
BMC Bioinformatics 2014, 15(345):1-16, October 2014. |
Journal Paper (without talk) |
 |
Manuela Waldner , Stefan Bruckner , Stefan Bruckner , Ivan Viola , Ivan Viola
Graphical Histories of Information Foraging
In Proceedings of the 8th Nordic Conference on Human-Computer Interaction: Fun, Fast, Foundational , pages 295-304. October 2014.
[ paper] paper] |
Conference Paper |
 |
Åsmund Birkeland, Veronika Solteszova, Dieter Hönigmann, Odd Helge Gilja, Svein Brekke, Timo Ropinski, Ivan Viola
The Ultrasound Visualization Pipeline
In Scientific Visualization, pages 283-303, September 2014
[ paper] paper] |
Article in a Book |
 |
Daniel Gehrer
CellUnity an Interactive Tool for Illustrative Visualization of Molecular Reactions
[ Bakk Thesis] Bakk Thesis] |
Bachelor Thesis |


 paper
paper
 Microtubule Catastrophe
Microtubule Catastrophe
 image
image Master Thesis
Master Thesis
 image
image Master Thesis
Master Thesis
 paper
paper
 image
image Master Thesis
Master Thesis

 Bachelor Thesis
Bachelor Thesis
 Bachelor Thesis
Bachelor Thesis
 paper
paper
 thesis
thesis
 Paper
Paper
 image
image paper
paper
 paper
paper paper
paper


 Paper
Paper Preview Movie
Preview Movie
 paper
paper video
video


 image
image
 Thesis
Thesis
 Paper
Paper
 paper
paper
 thesis
thesis Thesis
Thesis
 paper
paper
 paper
paper
 paper
paper
 paper
paper
 Paper
Paper
 paper
paper

 Paper
Paper
 Paper
Paper
 paper
paper screenshot
screenshot
 Paper
Paper

 paper
paper Preview video
Preview video Submission video
Submission video
 thesis
thesis


 paper
paper
 paper
paper
 Bakk Thesis
Bakk Thesis